Structure of Prototypic Peptide Transporter DtpA from E. coli in Complex with Valganciclovir Provides Insights into Drug Binding of Human PepT1.
Ural-Blimke, Y., Flayhan, A., Strauss, J., Rantos, V., Bartels, K., Nielsen, R., Pardon, E., Steyaert, J., Kosinski, J., Quistgaard, E.M., Low, C.(2019) J Am Chem Soc 141: 2404-2412
- PubMed: 30644743
- DOI: https://doi.org/10.1021/jacs.8b11343
- Primary Citation of Related Structures:
6GS1, 6GS4, 6GS7 - PubMed Abstract:
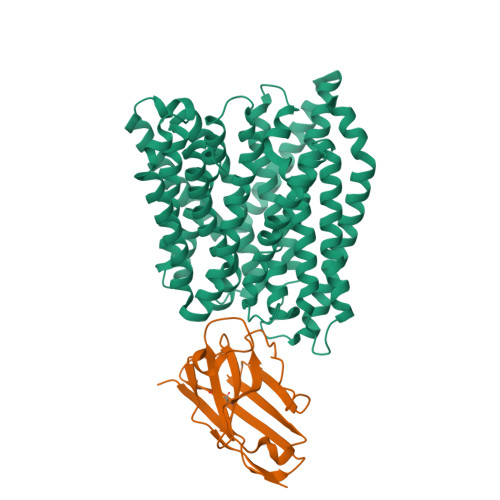
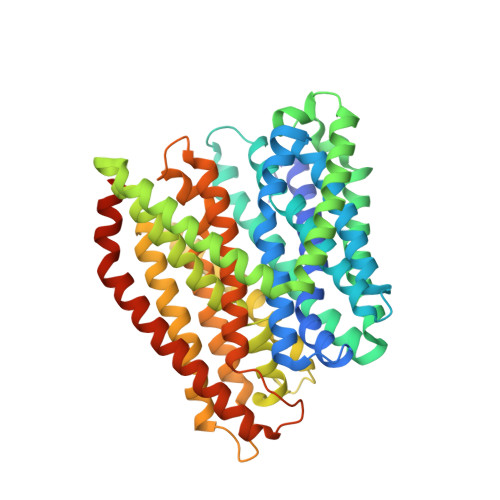
Members of the solute carrier 15 family (SLC15) transport di- and tripeptides as well as peptidomimetic drugs across the cell membrane. Structures of bacterial homologues have provided valuable information on the binding and transport of their natural substrates, but many do not transport medically relevant drugs. In contrast, a homologue from Escherichia coli, DtpA (dipeptide and tripeptide permease), shows a high similarity to human PepT1 (SLC15A1) in terms of ligand selectivity and transports a similar set of drugs. Here, we present the crystal structure of DtpA in ligand-free form (at 3.30 Å resolution) and in complex with the antiviral prodrug valganciclovir (at 2.65 Å resolution) supported by biochemical data. We show that valganciclovir unexpectedly binds with the ganciclovir moiety mimicking the N-terminal residue of a canonical peptide substrate. On the basis of a homology model we argue that this binding mode also applies to the human PepT1 transporter. Our results provide new insights into the binding mode of prodrugs and will assist the rational design of drugs with improved absorption rates.
Organizational Affiliation:
Centre for Structural Systems Biology (CSSB) , DESY and European Molecular Biology Laboratory Hamburg , Notkestrasse 85 , D-22607 Hamburg , Germany.