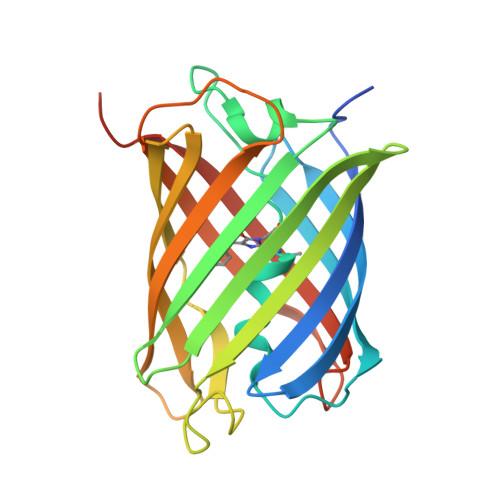
Structural insights into the binding of nanobodies LaM2 and LaM4 to the red fluorescent protein mCherry.
Wang, Z., Li, L., Hu, R., Zhong, P., Zhang, Y., Cheng, S., Jiang, H., Liu, R., Ding, Y.(2021) Protein Sci 30: 2298-2309
- PubMed: 34562299
- DOI: https://doi.org/10.1002/pro.4194
- Primary Citation of Related Structures:
6IR1, 6IR2 - PubMed Abstract:
Red fluorescent proteins (RFPs) are powerful tools used in molecular biology research. Although RFP can be easily monitored in vivo, manipulation of RFP by suitable nanobodies binding to different epitopes of RFP is still desired. Thus, it is crucial to obtain structural information on how the different nanobodies interact with RFP. Here, we determined the crystal structures of the LaM2-mCherry and LaM4-mCherry complexes at 1.4 and 1.9 Å resolution. Our results showed that LaM2 binds to the side of the mCherry β-barrel, while LaM4 binds to the bottom of the β-barrel. The distinct binding sites of LaM2 and LaM4 were further verified by isothermal titration calorimetry, fluorescence-based size exclusion chromatography, and dynamic light scattering assays. Mutation of the residues at the LaM2 or LaM4 binding interface to mCherry significantly decreased the binding affinity of the nanobody to mCherry. Our results also showed that LaM2 and LaM4 can bind to mCherry simultaneously, which is crucial for recruiting multiple operation elements to the RFP. The binding of LaM2 or LaM4 did not significantly change the chromophore environment of mCherry, which is important for fluorescence quantification assays, while several GFP nanobodies significantly altered the fluorescence. Our results provide atomic resolution interaction information on the binding of nanobodies LaM2 and LaM4 with mCherry, which is important for developing detection and manipulation methods for RFP-based biotechnology.
Organizational Affiliation:
School of Life Sciences, Fudan University, Shanghai, China.