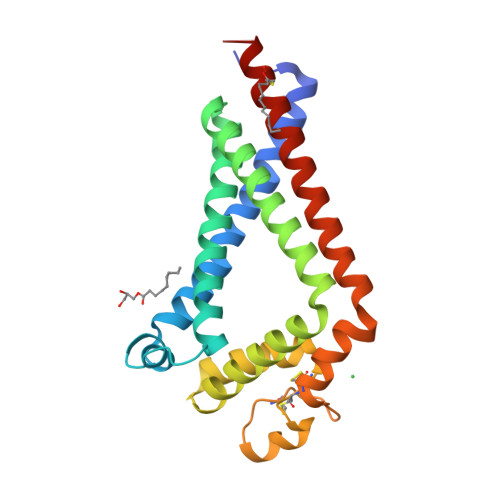
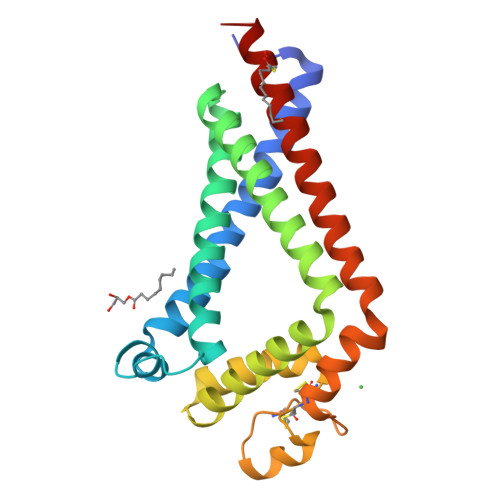
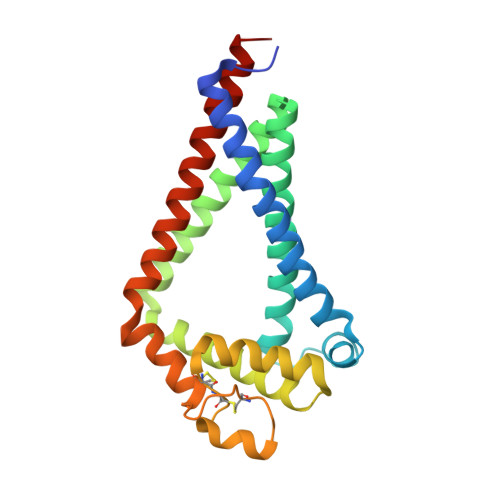
Structural insights into tetraspanin CD9 function.
Umeda, R., Satouh, Y., Takemoto, M., Nakada-Nakura, Y., Liu, K., Yokoyama, T., Shirouzu, M., Iwata, S., Nomura, N., Sato, K., Ikawa, M., Nishizawa, T., Nureki, O.(2020) Nat Commun 11: 1606-1606
- PubMed: 32231207
- DOI: https://doi.org/10.1038/s41467-020-15459-7
- Primary Citation of Related Structures:
6K4J - PubMed Abstract:
Tetraspanins play critical roles in various physiological processes, ranging from cell adhesion to virus infection. The members of the tetraspanin family have four membrane-spanning domains and short and large extracellular loops, and associate with a broad range of other functional proteins to exert cellular functions. Here we report the crystal structure of CD9 and the cryo-electron microscopic structure of CD9 in complex with its single membrane-spanning partner protein, EWI-2. The reversed cone-like molecular shape of CD9 generates membrane curvature in the crystalline lipid layers, which explains the CD9 localization in regions with high membrane curvature and its implications in membrane remodeling. The molecular interaction between CD9 and EWI-2 is mainly mediated through the small residues in the transmembrane region and protein/lipid interactions, whereas the fertilization assay revealed the critical involvement of the LEL region in the sperm-egg fusion, indicating the different dependency of each binding domain for other partner proteins.
Organizational Affiliation:
Department of Biological Sciences Graduate School of Science, The University of Tokyo, 7-3-1 Hongo, Bunkyo-ku, Tokyo, Japan.