A protein scaffold, engineered SPINK2, for generation of inhibitors with high affinity and specificity against target proteases.
Nishimiya, D., Kawaguchi, Y., Kodama, S., Nasu, H., Yano, H., Yamaguchi, A., Tamura, M., Hashimoto, R.(2019) Sci Rep 9: 11436-11436
- PubMed: 31391482
- DOI: https://doi.org/10.1038/s41598-019-47615-5
- Primary Citation of Related Structures:
6KBR - PubMed Abstract:
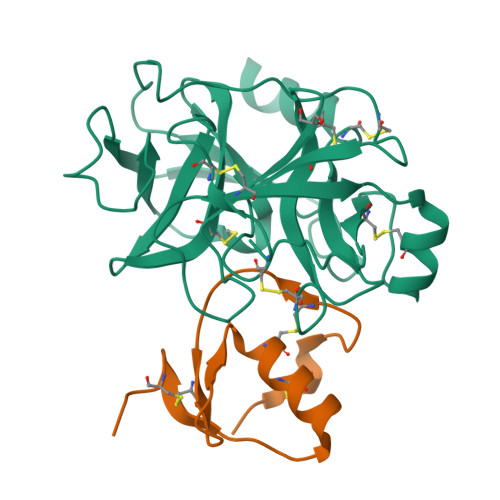
Proteases are one of attractive therapeutic targets to play key roles in pharmacological action. There are many protease inhibitors in nature, and most of them structurally have cystine knot motifs. Their structures are favorable for recognition of active pockets of proteases, leading to the potent inhibition. However, they also have drawbacks, such as broad cross-reactivity, on the therapeutic application. To create therapeutic proteins derived from a disulfide-rich scaffold, we selected human serine protease inhibitor Kazal type 2 (SPINK2) through a scaffold screening, as a protein scaffold with requirements for therapeutic proteins. We then constructed a diverse library of the engineered SPINK2 by introducing random mutations into its flexible loop region with the designed method. By phage panning against four serine proteases, we isolated potent inhibitors against each target with picomolar K D and sub-nanomolar K i values. Also, they exhibited the desired specificities against target proteases without inhibiting non-target proteases. The crystal structure of kallikrein related peptidase 4 (KLK4)-engineered SPINK2 complex revealed the interface with extensive conformational complementarity. Our study demonstrates that engineered SPINK2 can serve as a scaffold to generate therapeutic molecules against target proteins with groove structures.
Organizational Affiliation:
DAIICHI SANKYO CO., LTD., Biologics Division, Modality Research Laboratories, 1-2-58, Hiromachi, Shinagawa-ku, Tokyo, 140-8710, Japan. nishimiya.daisuke.tc@daiichisankyo.co.jp.