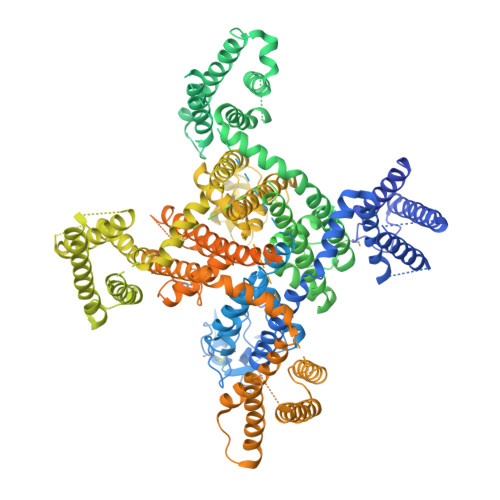
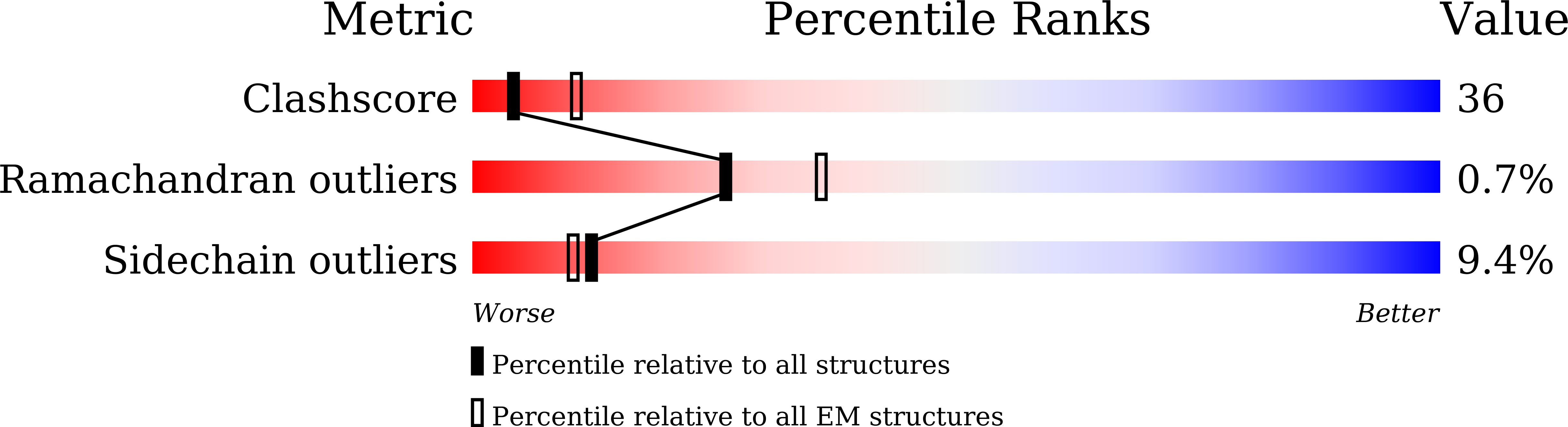Cryo-EM structures of apo and antagonist-bound human Cav3.1.
Zhao, Y., Huang, G., Wu, Q., Wu, K., Li, R., Lei, J., Pan, X., Yan, N.(2019) Nature 576: 492-497
- PubMed: 31766050
- DOI: https://doi.org/10.1038/s41586-019-1801-3
- Primary Citation of Related Structures:
6KZO, 6KZP - PubMed Abstract:
Among the ten subtypes of mammalian voltage-gated calcium (Ca v ) channels, Ca v 3.1-Ca v 3.3 constitute the T-type, or the low-voltage-activated, subfamily, the abnormal activities of which are associated with epilepsy, psychiatric disorders and pain 1-5 . Here we report the cryo-electron microscopy structures of human Ca v 3.1 alone and in complex with a highly Ca v 3-selective blocker, Z944 6,7 , at resolutions of 3.3 Å and 3.1 Å, respectively. The arch-shaped Z944 molecule reclines in the central cavity of the pore domain, with the wide end inserting into the fenestration on the interface between repeats II and III, and the narrow end hanging above the intracellular gate like a plug. The structures provide the framework for comparative investigation of the distinct channel properties of different Ca v subfamilies.
Organizational Affiliation:
Key Laboratory of Structural Biology of Zhejiang Province, School of Life Sciences, Westlake University, Hangzhou, China.