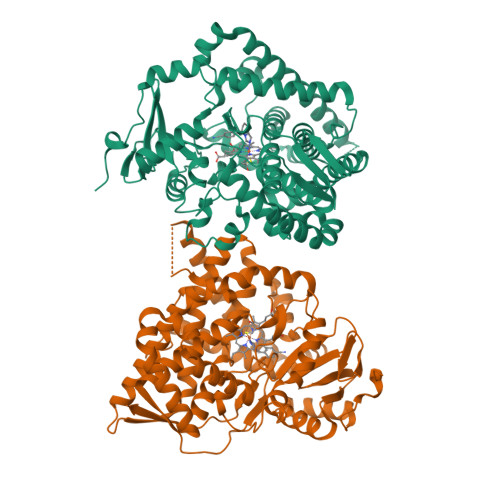
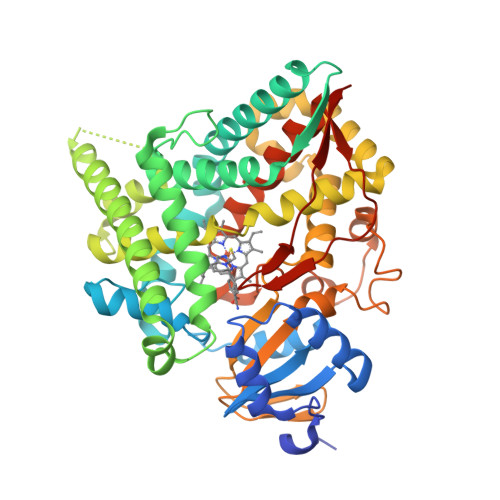
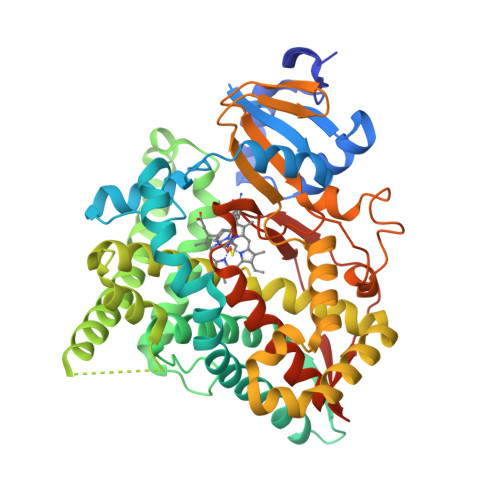
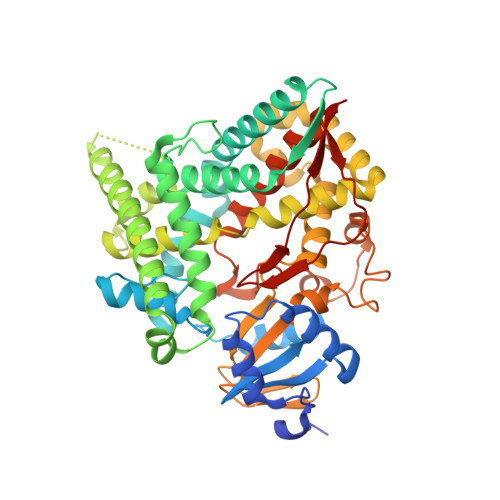
Structure of human cortisol-producing cytochrome P450 11B1 bound to the breast cancer drug fadrozole provides insights for drug design.
Brixius-Anderko, S., Scott, E.E.(2019) J Biological Chem 294: 453-460
- PubMed: 30425102
- DOI: https://doi.org/10.1074/jbc.RA118.006214
- Primary Citation of Related Structures:
6M7X - PubMed Abstract:
Human cytochrome P450 11B1 (CYP11B1) is responsible for the final step generating the steroid hormone cortisol, which controls stress and immune responses and glucose homeostasis. CYP11B1 is a promising drug target to manage Cushing's disease, a disorder arising from excessive cortisol production. However, the design of selective inhibitors has been hampered because structural information for CYP11B1 is unavailable and the enzyme has high amino acid sequence identity (93%) to a closely related enzyme, the aldosterone-producing CYP11B2. Here we report the X-ray crystal structure of human CYP11B1 (at 2.1 Å resolution) in complex with fadrozole, a racemic compound normally used to treat breast cancer by inhibiting estrogen-producing CYP19A1. Comparison of fadrozole-bound CYP11B1 with fadrozole-bound CYP11B2 revealed that despite conservation of the active-site residues, the overall structures and active sites had structural rearrangements consistent with distinct protein functions and inhibition. Whereas fadrozole binds to both CYP11B enzymes by coordinating the heme iron, CYP11B2 binds to the R enantiomer of fadrozole, and CYP11B1 binds to the S enantiomer, each with distinct orientations and interactions. These results provide insights into the cross-reactivity of drugs across multiple steroidogenic cytochrome P450 enzymes, provide a structural basis for understanding human steroidogenesis, and pave the way for the design of more selective inhibitors of each human CYP11B enzyme.
Organizational Affiliation:
From the Departments of Medicinal Chemistry and.