The structure of the extracellular domains of human interleukin 11 alpha receptor reveals mechanisms of cytokine engagement.
Metcalfe, R.D., Aizel, K., Zlatic, C.O., Nguyen, P.M., Morton, C.J., Lio, D.S., Cheng, H.C., Dobson, R.C.J., Parker, M.W., Gooley, P.R., Putoczki, T.L., Griffin, M.D.W.(2020) J Biological Chem 295: 8285-8301
- PubMed: 32332100
- DOI: https://doi.org/10.1074/jbc.RA119.012351
- Primary Citation of Related Structures:
6O4O, 6O4P - PubMed Abstract:
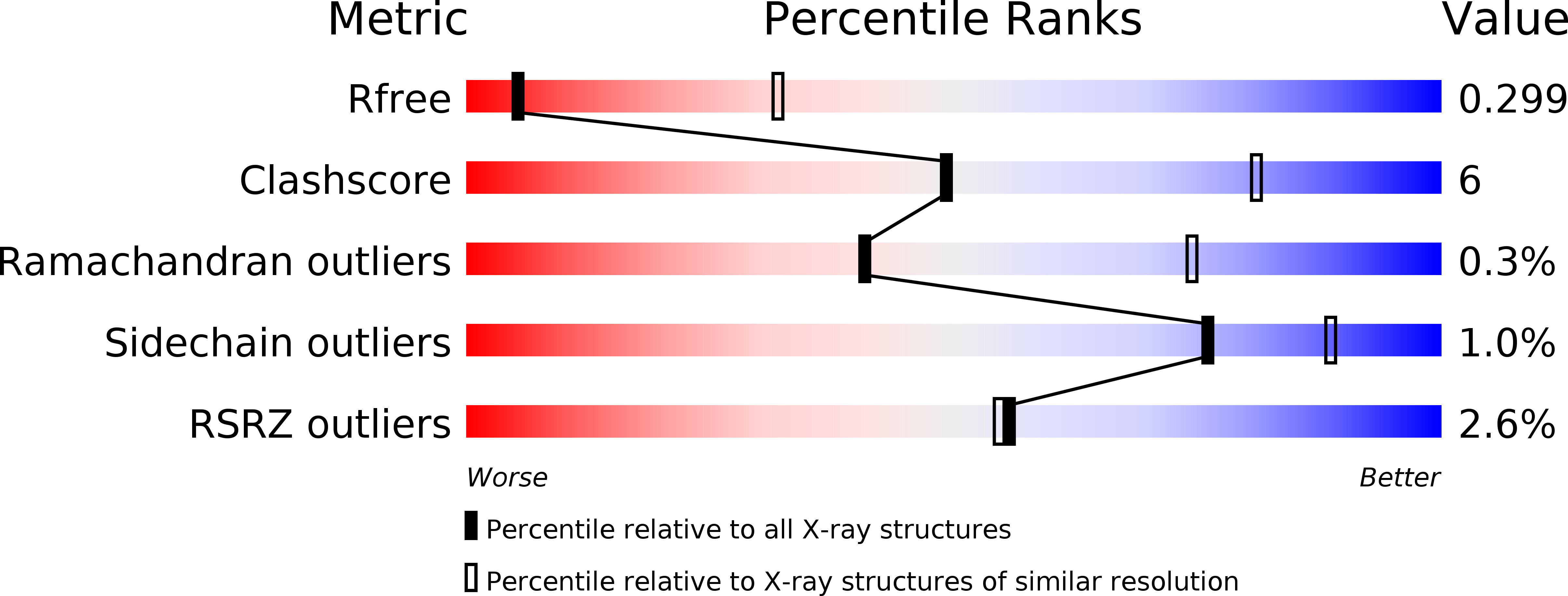
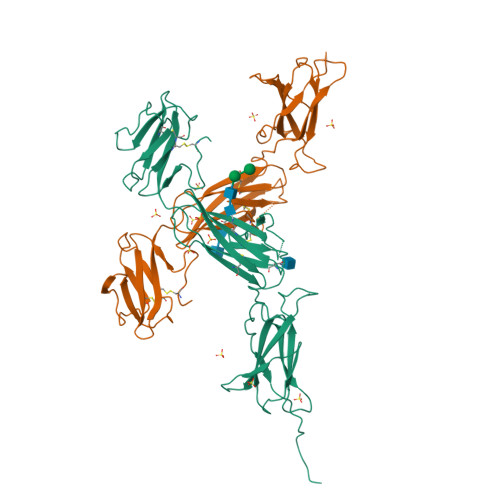
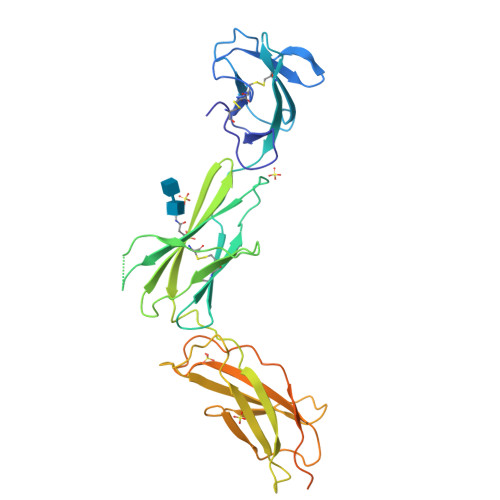
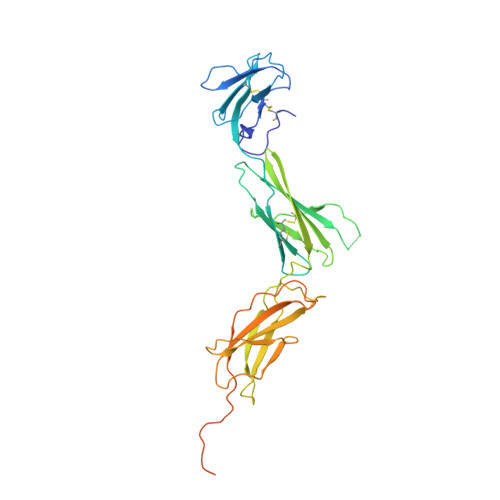
Interleukin (IL) 11 activates multiple intracellular signaling pathways by forming a complex with its cell surface α-receptor, IL-11Rα, and the β-subunit receptor, gp130. Dysregulated IL-11 signaling has been implicated in several diseases, including some cancers and fibrosis. Mutations in IL-11Rα that reduce signaling are also associated with hereditary cranial malformations. Here we present the first crystal structure of the extracellular domains of human IL-11Rα and a structure of human IL-11 that reveals previously unresolved detail. Disease-associated mutations in IL-11Rα are generally distal to putative ligand-binding sites. Molecular dynamics simulations showed that specific mutations destabilize IL-11Rα and may have indirect effects on the cytokine-binding region. We show that IL-11 and IL-11Rα form a 1:1 complex with nanomolar affinity and present a model of the complex. Our results suggest that the thermodynamic and structural mechanisms of complex formation between IL-11 and IL-11Rα differ substantially from those previously reported for similar cytokines. This work reveals key determinants of the engagement of IL-11 by IL-11Rα that may be exploited in the development of strategies to modulate formation of the IL-11-IL-11Rα complex.
Organizational Affiliation:
Department of Biochemistry and Molecular Biology, Bio21 Molecular Science and Biotechnology Institute.