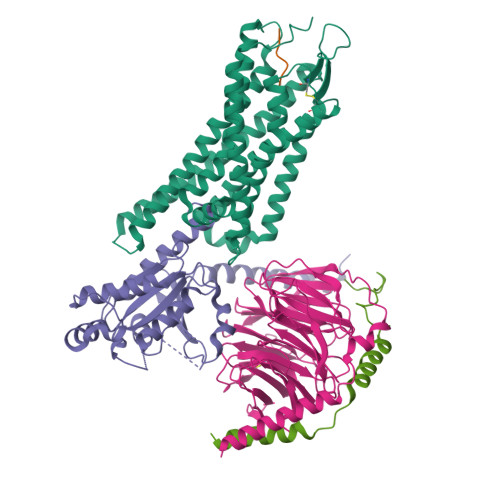
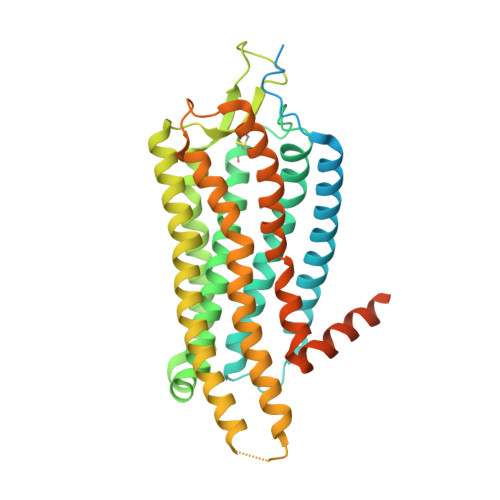
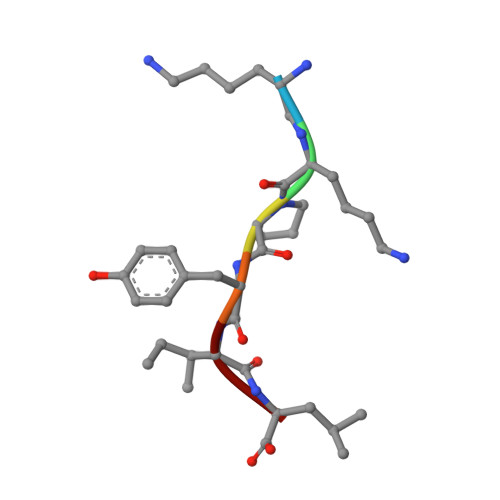
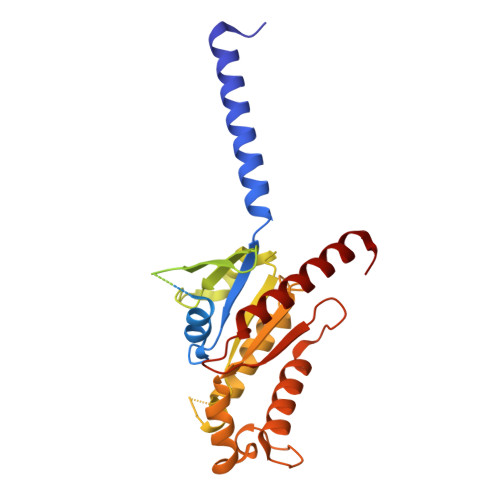
Conformational transitions of a neurotensin receptor 1-Gi1complex.
Kato, H.E., Zhang, Y., Hu, H., Suomivuori, C.M., Kadji, F.M.N., Aoki, J., Krishna Kumar, K., Fonseca, R., Hilger, D., Huang, W., Latorraca, N.R., Inoue, A., Dror, R.O., Kobilka, B.K., Skiniotis, G.(2019) Nature 572: 80-85
- PubMed: 31243364
- DOI: https://doi.org/10.1038/s41586-019-1337-6
- Primary Citation of Related Structures:
6OS9, 6OSA - PubMed Abstract:
Neurotensin receptor 1 (NTSR1) is a G-protein-coupled receptor (GPCR) that engages multiple subtypes of G protein, and is involved in the regulation of blood pressure, body temperature, weight and the response to pain. Here we present structures of human NTSR1 in complex with the agonist JMV449 and the heterotrimeric G i1 protein, at a resolution of 3 Å. We identify two conformations: a canonical-state complex that is similar to recently reported GPCR-G i/o complexes (in which the nucleotide-binding pocket adopts more flexible conformations that may facilitate nucleotide exchange), and a non-canonical state in which the G protein is rotated by about 45 degrees relative to the receptor and exhibits a more rigid nucleotide-binding pocket. In the non-canonical state, NTSR1 exhibits features of both active and inactive conformations, which suggests that the structure may represent an intermediate form along the activation pathway of G proteins. This structural information, complemented by molecular dynamics simulations and functional studies, provides insights into the complex process of G-protein activation.
Organizational Affiliation:
Department of Molecular and Cellular Physiology, Stanford University School of Medicine, Stanford, CA, USA.