Novel, Self-Assembling Dimeric Inhibitors of Human beta Tryptase.
Giardina, S.F., Werner, D.S., Pingle, M., Feinberg, P.B., Foreman, K.W., Bergstrom, D.E., Arnold, L.D., Barany, F.(2020) J Med Chem 63: 3004-3027
- PubMed: 32057241
- DOI: https://doi.org/10.1021/acs.jmedchem.9b01689
- Primary Citation of Related Structures:
6P0P - PubMed Abstract:
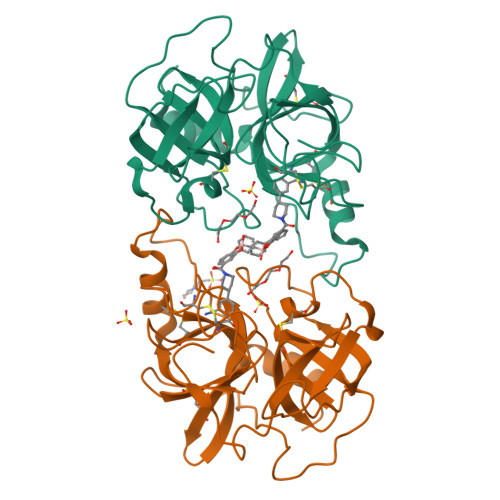
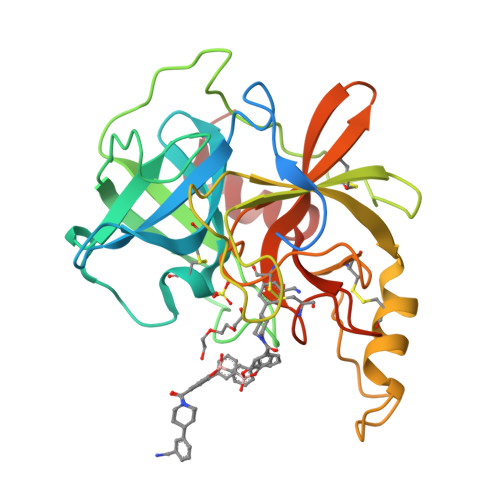
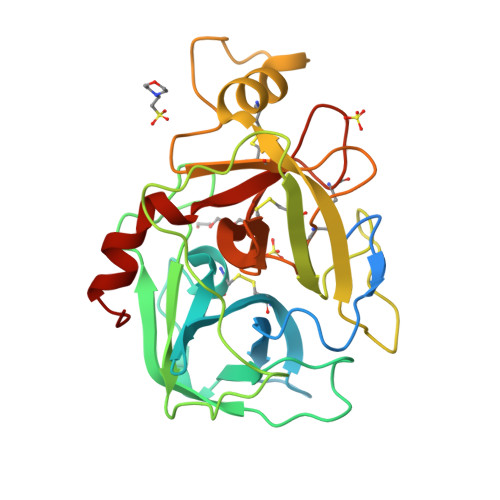
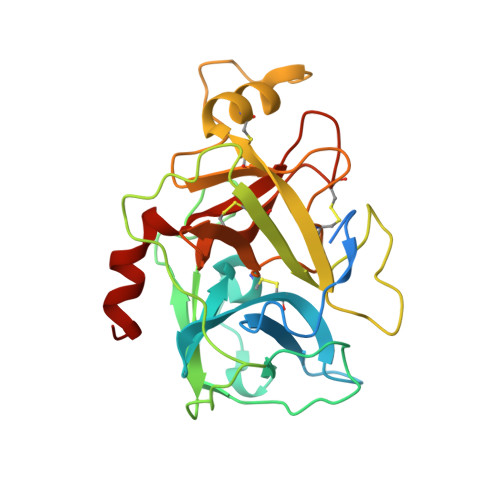
β-Tryptase, a homotetrameric serine protease, has four identical active sites facing a central pore, presenting an optimized setting for the rational design of bivalent inhibitors that bridge two adjacent sites. Using diol, hydroxymethyl phenols or benzoyl methyl hydroxamates, and boronic acid chemistries to reversibly join two [3-(1-acylpiperidin-4-yl)phenyl]methanamine core ligands, we have successfully produced a series of self-assembling heterodimeric inhibitors. These heterodimeric tryptase inhibitors demonstrate superior activity compared to monomeric modes of inhibition. X-ray crystallography validated the dimeric mechanism of inhibition, and compounds demonstrated high selectivity against related proteases, good target engagement, and tryptase inhibition in HMC1 xenograft models. Screening 3872 possible combinations from 44 boronic acid and 88 diol derivatives revealed several combinations that produced nanomolar inhibition, and seven unique pairs produced greater than 100-fold improvement in potency over monomeric inhibition. These heterodimeric tryptase inhibitors demonstrate the power of target-driven combinatorial chemistry to deliver bivalent drugs in a small molecule form.
Organizational Affiliation:
Department of Microbiology and Immunology, Weill Cornell Medicine, 1300 York Avenue, Box 62, New York, New York 10065, United States.