A conformation-specific ON-switch for controlling CAR T cells with an orally available drug.
Zajc, C.U., Dobersberger, M., Schaffner, I., Mlynek, G., Puhringer, D., Salzer, B., Djinovic-Carugo, K., Steinberger, P., De Sousa Linhares, A., Yang, N.J., Obinger, C., Holter, W., Traxlmayr, M.W., Lehner, M.(2020) Proc Natl Acad Sci U S A 117: 14926-14935
- PubMed: 32554495
- DOI: https://doi.org/10.1073/pnas.1911154117
- Primary Citation of Related Structures:
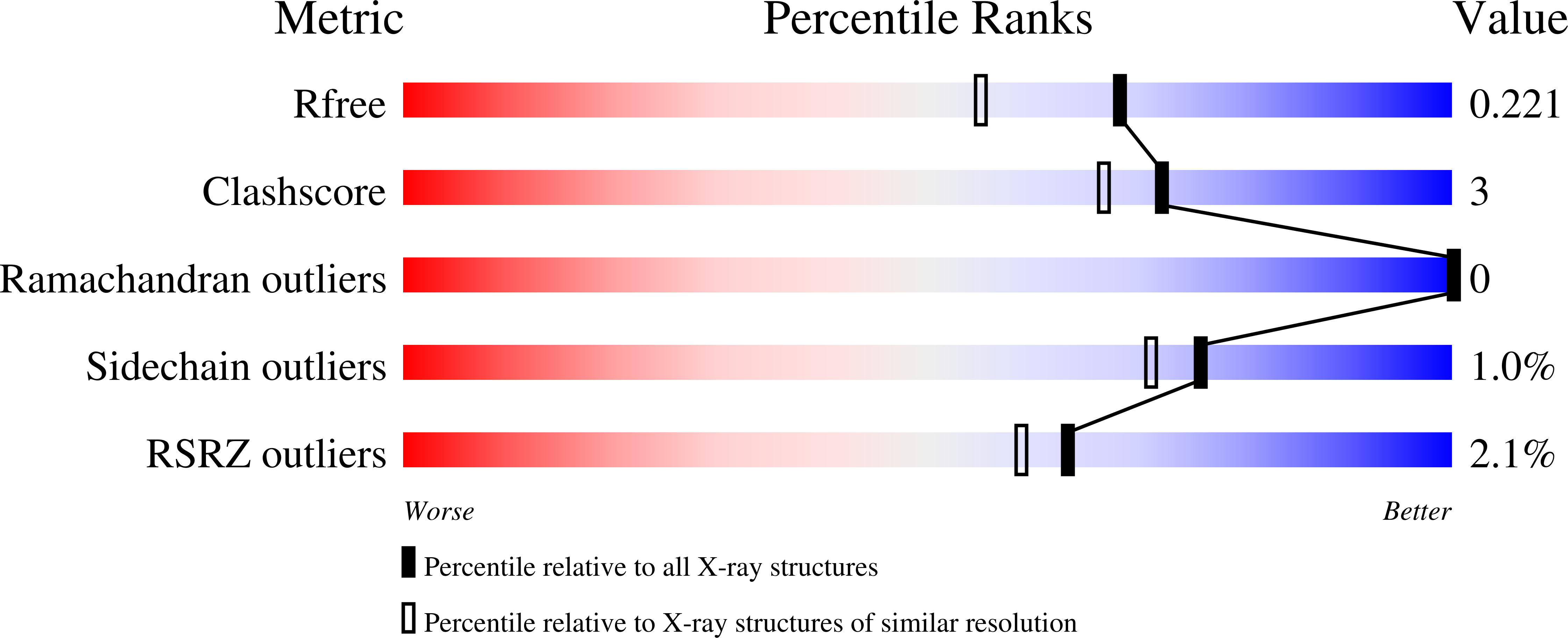
6QBA - PubMed Abstract:
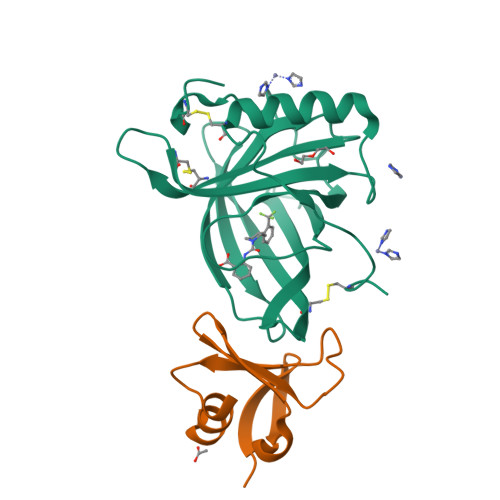
Molecular ON-switches in which a chemical compound induces protein-protein interactions can allow cellular function to be controlled with small molecules. ON-switches based on clinically applicable compounds and human proteins would greatly facilitate their therapeutic use. Here, we developed an ON-switch system in which the human retinol binding protein 4 (hRBP4) of the lipocalin family interacts with engineered hRBP4 binders in a small molecule-dependent manner. Two different protein scaffolds were engineered to bind to hRBP4 when loaded with the orally available small molecule A1120. The crystal structure of an assembled ON-switch shows that the engineered binder specifically recognizes the conformational changes induced by A1120 in two loop regions of hRBP4. We demonstrate that this conformation-specific ON-switch is highly dependent on the presence of A1120, as demonstrated by an ∼500-fold increase in affinity upon addition of the small molecule drug. Furthermore, the ON-switch successfully regulated the activity of primary human CAR T cells in vitro. We anticipate that lipocalin-based ON-switches have the potential to be broadly applied for the safe pharmacological control of cellular therapeutics.
Organizational Affiliation:
St. Anna Children's Cancer Research Institute (CCRI), 1090 Vienna, Austria.