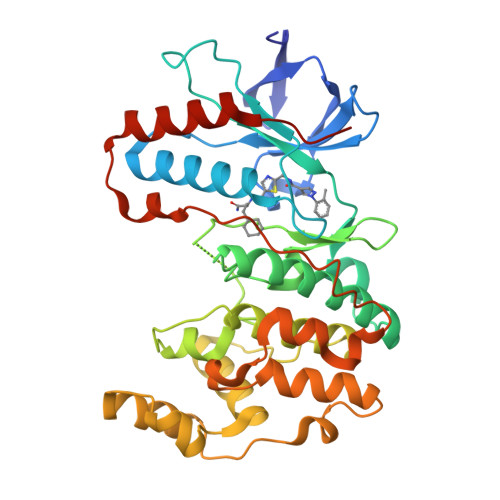
Fast Iterative Synthetic Approach toward Identification of Novel Highly Selective p38 MAP Kinase Inhibitors.
Rohm, S., Berger, B.T., Schroder, M., Chaikuad, A., Winkel, R., Hekking, K.F.W., Benningshof, J.J.C., Muller, G., Tesch, R., Kudolo, M., Forster, M., Laufer, S., Knapp, S.(2019) J Med Chem 62: 10757-10782
- PubMed: 31702918
- DOI: https://doi.org/10.1021/acs.jmedchem.9b01227
- Primary Citation of Related Structures:
6SFI, 6SFJ, 6SFK - PubMed Abstract:
p38 mitogen-activated protein kinases are key mediators of environmental stress response and are promising targets for treatment of inflammatory diseases and cancer. Numerous efforts have led to the discovery of several potent inhibitors; however, so far no highly selective type-II inhibitors have been reported. We previously identified VPC-00628 as a potent and selective type-II inhibitor of p38α/β with few off-targets. Here we analyzed the chemical building blocks of VPC-00628 that played a key role in achieving potency and selectivity through targeting an inactive state of the kinases induced by a unique folded P-loop conformation. Using a rapid, systematic combinatorial synthetic approach, we identified compound 93 ( SR-318 ) with excellent potency and selectivity for p38α/β, which potently inhibited the TNF-α release in whole blood. SR-318 therefore presents a potent and selective type-II inhibitor of p38α/β that can be used as a chemical probe for targeting this particular inactive state of these two p38 isoforms.
Organizational Affiliation:
Institute for Pharmaceutical Chemistry , Johann Wolfgang Goethe-University , Max-von-Laue-Str. 9 , D-60438 Frankfurt am Main , Germany.