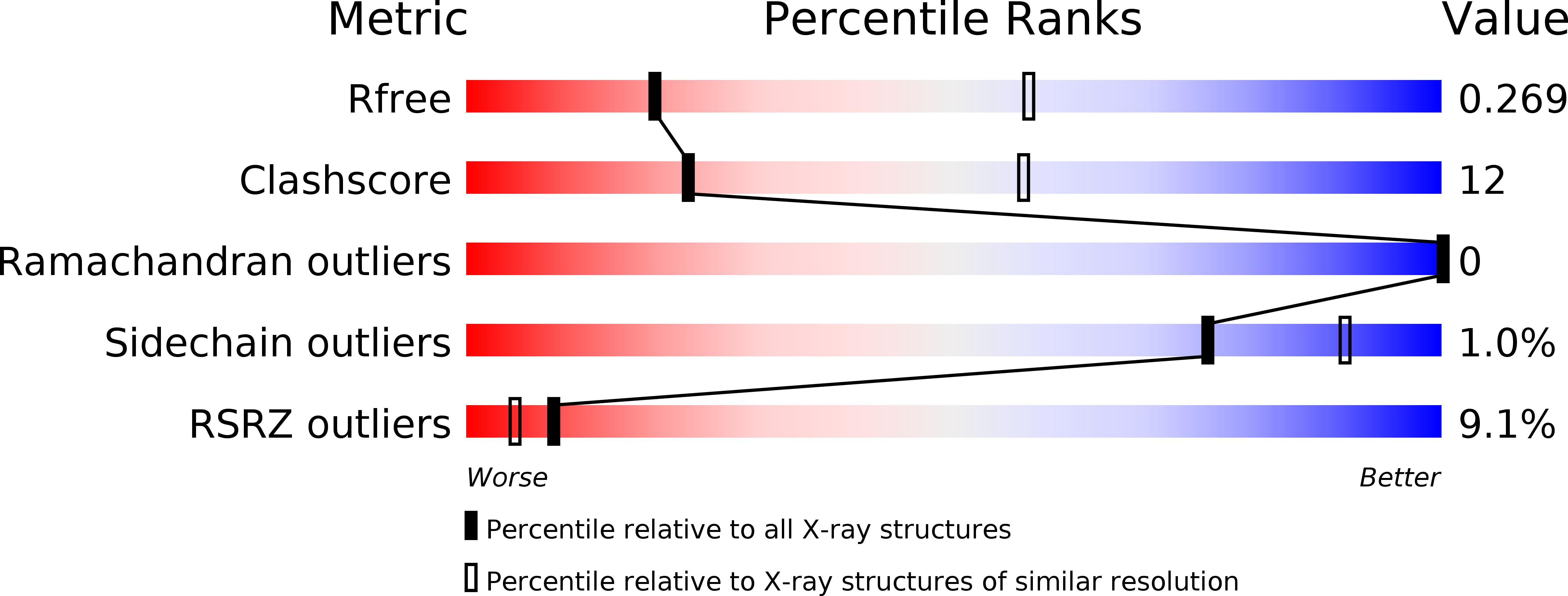
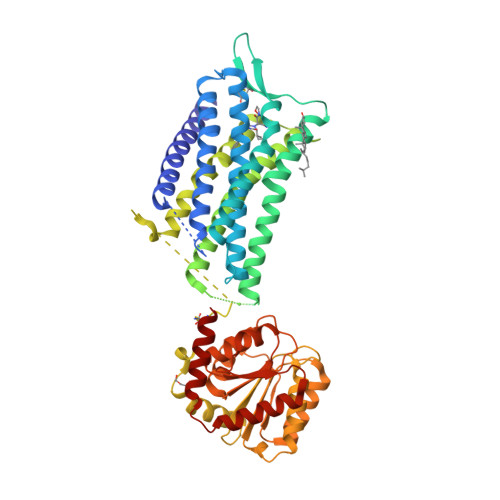
Crystal structure of the human oxytocin receptor.
Waltenspuhl, Y., Schoppe, J., Ehrenmann, J., Kummer, L., Pluckthun, A.(2020) Sci Adv 6: eabb5419-eabb5419
- PubMed: 32832646
- DOI: https://doi.org/10.1126/sciadv.abb5419
- Primary Citation of Related Structures:
6TPK - PubMed Abstract:
The peptide hormone oxytocin modulates socioemotional behavior and sexual reproduction via the centrally expressed oxytocin receptor (OTR) across several species. Here, we report the crystal structure of human OTR in complex with retosiban, a nonpeptidic antagonist developed as an oral drug for the prevention of preterm labor. Our structure reveals insights into the detailed interactions between the G protein-coupled receptor (GPCR) and an OTR-selective antagonist. The observation of an extrahelical cholesterol molecule, binding in an unexpected location between helices IV and V, provides a structural rationale for its allosteric effect and critical influence on OTR function. Furthermore, our structure in combination with experimental data allows the identification of a conserved neurohypophyseal receptor-specific coordination site for Mg 2+ that acts as potent, positive allosteric modulator for agonist binding. Together, these results further our molecular understanding of the oxytocin/vasopressin receptor family and will facilitate structure-guided development of new therapeutics.
Organizational Affiliation:
Department of Biochemistry, University of Zürich, Winterthurerstrasse 190, CH-8057 Zürich, Switzerland.