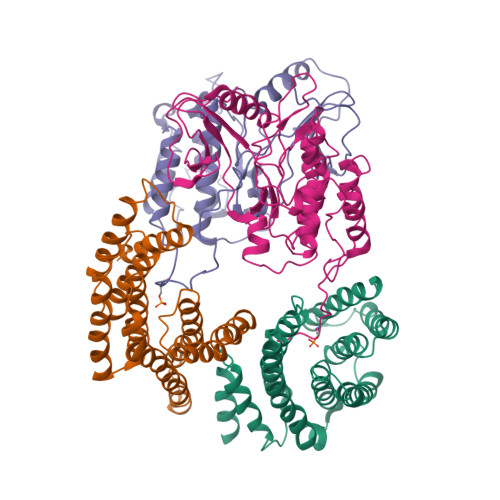
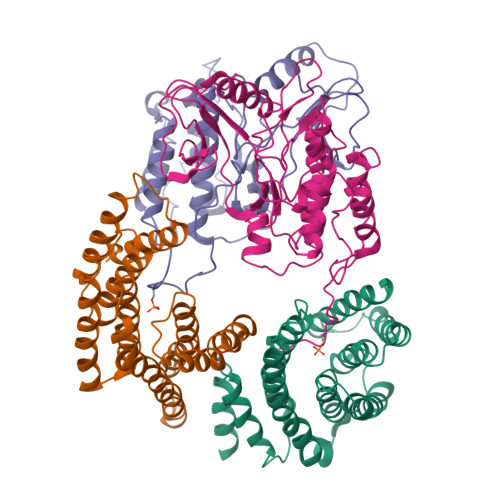
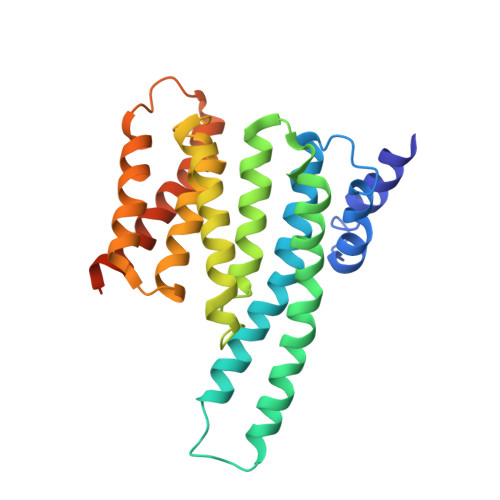
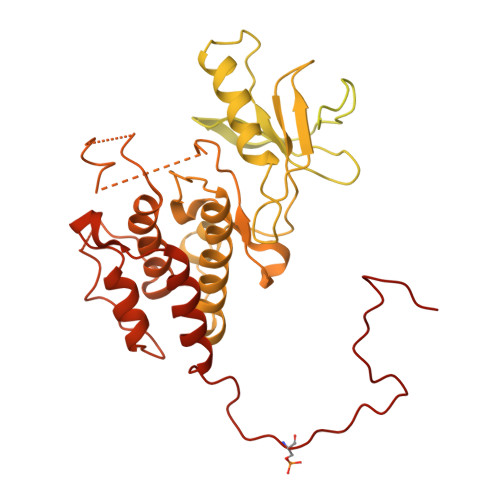
Cryo-EM structure of a dimeric B-Raf:14-3-3 complex reveals asymmetry in the active sites of B-Raf kinases.
Kondo, Y., Ognjenovic, J., Banerjee, S., Karandur, D., Merk, A., Kulhanek, K., Wong, K., Roose, J.P., Subramaniam, S., Kuriyan, J.(2019) Science 366: 109-115
- PubMed: 31604311
- DOI: https://doi.org/10.1126/science.aay0543
- Primary Citation of Related Structures:
6UAN - PubMed Abstract:
Raf kinases are important cancer drug targets. Paradoxically, many B-Raf inhibitors induce the activation of Raf kinases. Cryo-electron microscopy structural analysis of a phosphorylated B-Raf kinase domain dimer in complex with dimeric 14-3-3, at a resolution of ~3.9 angstroms, shows an asymmetric arrangement in which one kinase is in a canonical "active" conformation. The distal segment of the C-terminal tail of this kinase interacts with, and blocks, the active site of the cognate kinase in this asymmetric arrangement. Deletion of the C-terminal segment reduces Raf activity. The unexpected asymmetric quaternary architecture illustrates how the paradoxical activation of Raf by kinase inhibitors reflects an innate mechanism, with 14-3-3 facilitating inhibition of one kinase while maintaining activity of the other. Conformational modulation of these contacts may provide new opportunities for Raf inhibitor development.
Organizational Affiliation:
Department of Molecular and Cell Biology, University of California, Berkeley, Berkeley, CA 94720, USA.