Discovery of a First-in-Class Inhibitor of the PRMT5-Substrate Adaptor Interaction.
McKinney, D.C., McMillan, B.J., Ranaghan, M.J., Moroco, J.A., Brousseau, M., Mullin-Bernstein, Z., O'Keefe, M., McCarren, P., Mesleh, M.F., Mulvaney, K.M., Robinson, F., Singh, R., Bajrami, B., Wagner, F.F., Hilgraf, R., Drysdale, M.J., Campbell, A.J., Skepner, A., Timm, D.E., Porter, D., Kaushik, V.K., Sellers, W.R., Ianari, A.(2021) J Med Chem 64: 11148-11168
- PubMed: 34342224
- DOI: https://doi.org/10.1021/acs.jmedchem.1c00507
- Primary Citation of Related Structures:
6V0P, 7M05 - PubMed Abstract:
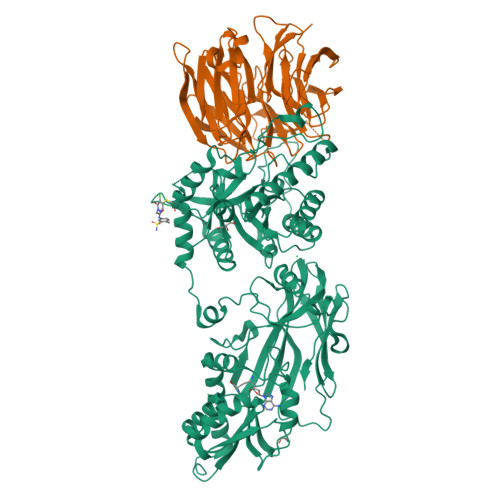
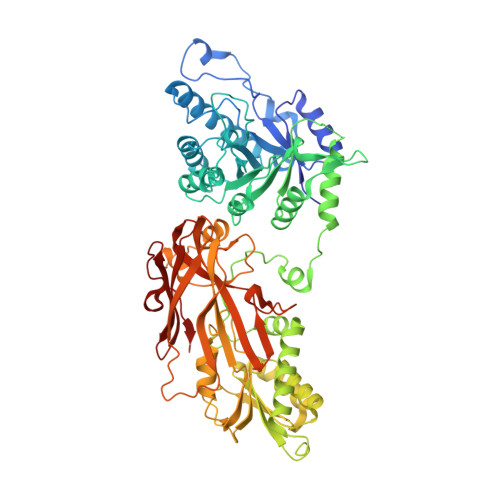
PRMT5 and its substrate adaptor proteins (SAPs), pICln and Riok1, are synthetic lethal dependencies in MTAP-deleted cancer cells. SAPs share a conserved PRMT5 binding motif (PBM) which mediates binding to a surface of PRMT5 distal to the catalytic site. This interaction is required for methylation of several PRMT5 substrates, including histone and spliceosome complexes. We screened for small molecule inhibitors of the PRMT5-PBM interaction and validated a compound series which binds to the PRMT5-PBM interface and directly inhibits binding of SAPs. Mode of action studies revealed the formation of a covalent bond between a halogenated pyridazinone group and cysteine 278 of PRMT5. Optimization of the starting hit produced a lead compound, BRD0639, which engages the target in cells, disrupts PRMT5-RIOK1 complexes, and reduces substrate methylation. BRD0639 is a first-in-class PBM-competitive inhibitor that can support studies of PBM-dependent PRMT5 activities and the development of novel PRMT5 inhibitors that selectively target these functions.
Organizational Affiliation:
Center for the Development of Therapeutics, The Broad Institute of MIT and Harvard, 415 Main Street, Cambridge, Massachusetts 02142, United States.