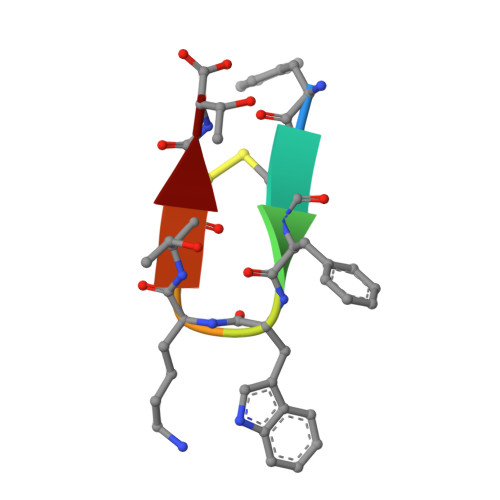
New perspectives in macromolecular powder diffraction using single-photon-counting strip detectors: high-resolution structure of the pharmaceutical peptide octreotide.
Spiliopoulou, M., Karavassili, F., Triandafillidis, D.P., Valmas, A., Fili, S., Kosinas, C., Barlos, K., Barlos, K.K., Morin, M., Reinle-Schmitt, M.L., Gozzo, F., Margiolaki, I.(2021) Acta Crystallogr A Found Adv 77: 186-195
- PubMed: 33944797
- DOI: https://doi.org/10.1107/S2053273321001698
- Primary Citation of Related Structures:
6VC1 - PubMed Abstract:
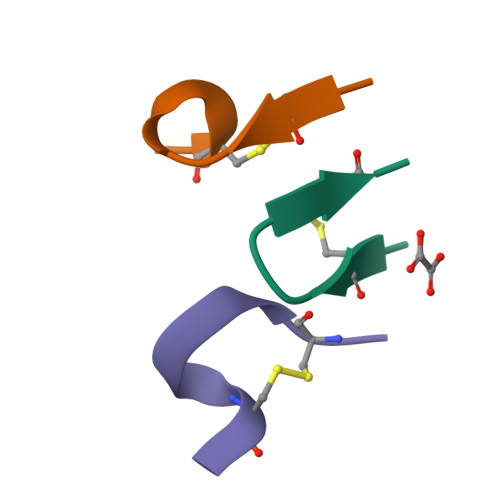
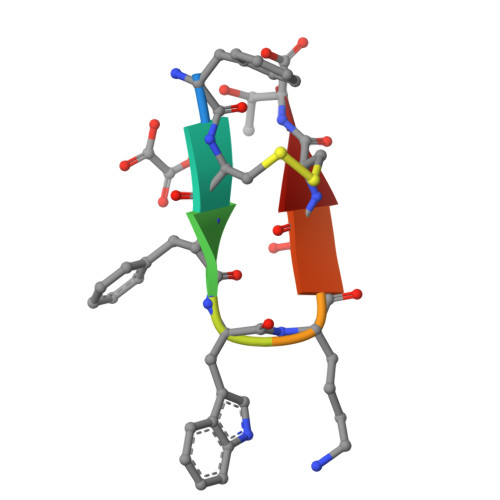
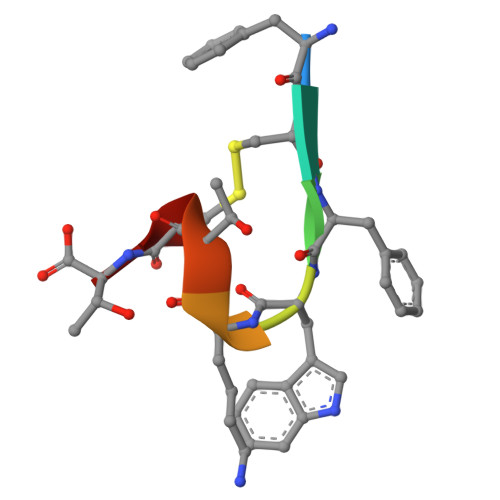
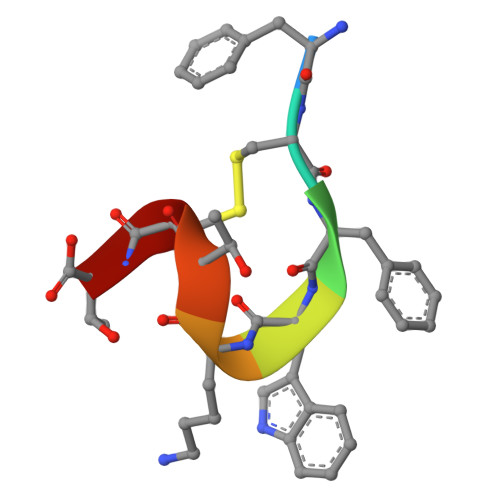
Advances in instrumentation, as well as the development of powerful crystallographic software have significantly facilitated the collection of high-resolution diffraction data and have made X-ray powder diffraction (XRPD) particularly useful for the extraction of structural information; this is true even for complex molecules, especially when combined with synchrotron radiation. In this study, in-line with past instrumental profile studies, an improved data collection strategy exploiting the MYTHEN II detector system together with significant beam focusing and tailored data collection options was introduced and optimized for protein samples at the Material Science beamline at the Swiss Light Source. Polycrystalline precipitates of octreotide, a somatostatin analog of particular pharmaceutical interest, were examined with this novel approach. XRPD experiments resulted in high angular and d-spacing (1.87 Å) resolution data, from which electron-density maps of enhanced quality were extracted, revealing the molecule's structural properties. Since microcrystalline precipitates represent a viable alternative for administration of therapeutic macromolecules, XRPD has been acknowledged as the most applicable tool for examining a wide spectrum of physicochemical properties of such materials and performing studies ranging from phase identification to complete structural characterization.
Organizational Affiliation:
Department of Biology, Section of Genetics, Cell Biology and Development, University of Patras, Patras, GR-26500, Greece.