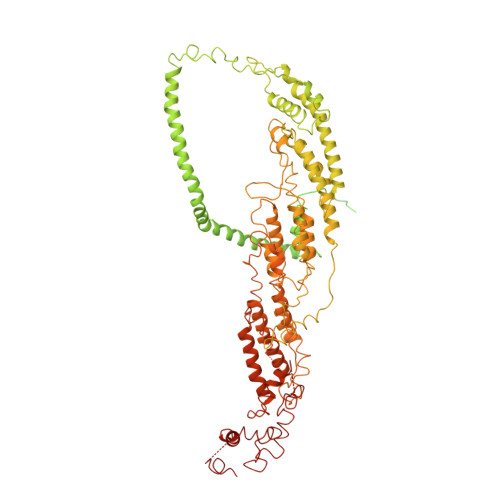
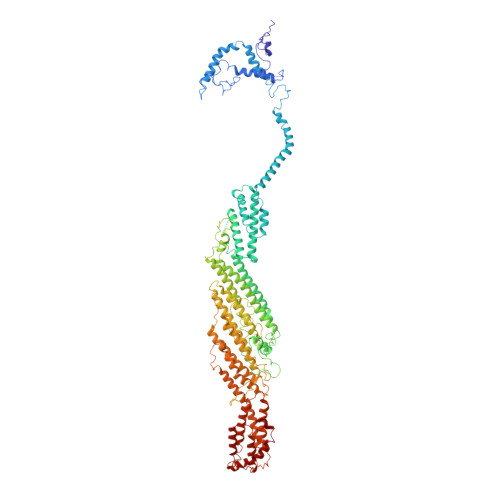
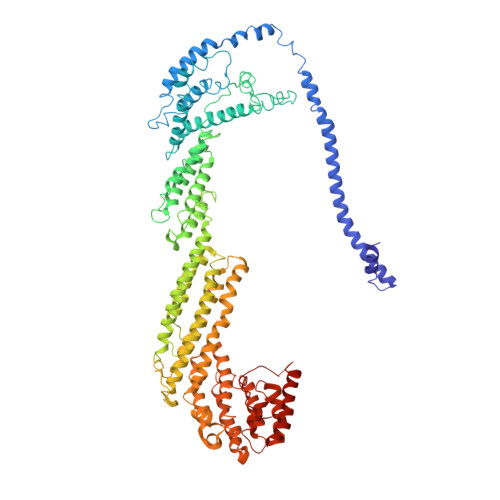
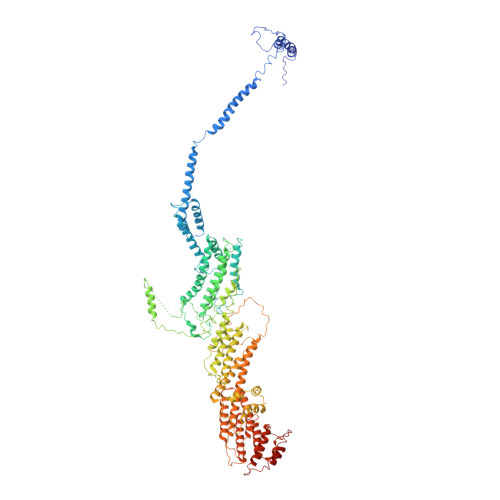
Exocyst structural changes associated with activation of tethering downstream of Rho/Cdc42 GTPases.
Rossi, G., Lepore, D., Kenner, L., Czuchra, A.B., Plooster, M., Frost, A., Munson, M., Brennwald, P.(2020) J Cell Biol 219
- PubMed: 31904797
- DOI: https://doi.org/10.1083/jcb.201904161
- Primary Citation of Related Structures:
6VKL - PubMed Abstract:
The exocyst complex plays a critical role in determining both temporal and spatial dynamics of exocytic vesicle tethering and fusion with the plasma membrane. However, the mechanism by which the exocyst functions and how it is regulated remain poorly understood. Here we describe a novel biochemical assay for the examination of exocyst function in vesicle tethering. Importantly, the assay is stimulated by gain-of-function mutations in the Exo70 component of the exocyst, selected for their ability to bypass Rho/Cdc42 activation in vivo. Single-particle electron microscopy and 3D reconstructions of negatively stained exocyst complexes reveal a structural change in the mutant exocyst that exposes a binding site for the v-SNARE. We demonstrate a v-SNARE requirement in our tethering assay and increased v-SNARE binding to exocyst gain-of-function complexes. Together, these data suggest an allosteric mechanism for activation involving a conformational change in one subunit of the complex, which is relayed through the complex to regulate its biochemical activity in vitro, as well as overall function in vivo.
Organizational Affiliation:
Department of Cell Biology and Physiology, University of North Carolina School of Medicine, Chapel Hill, NC.