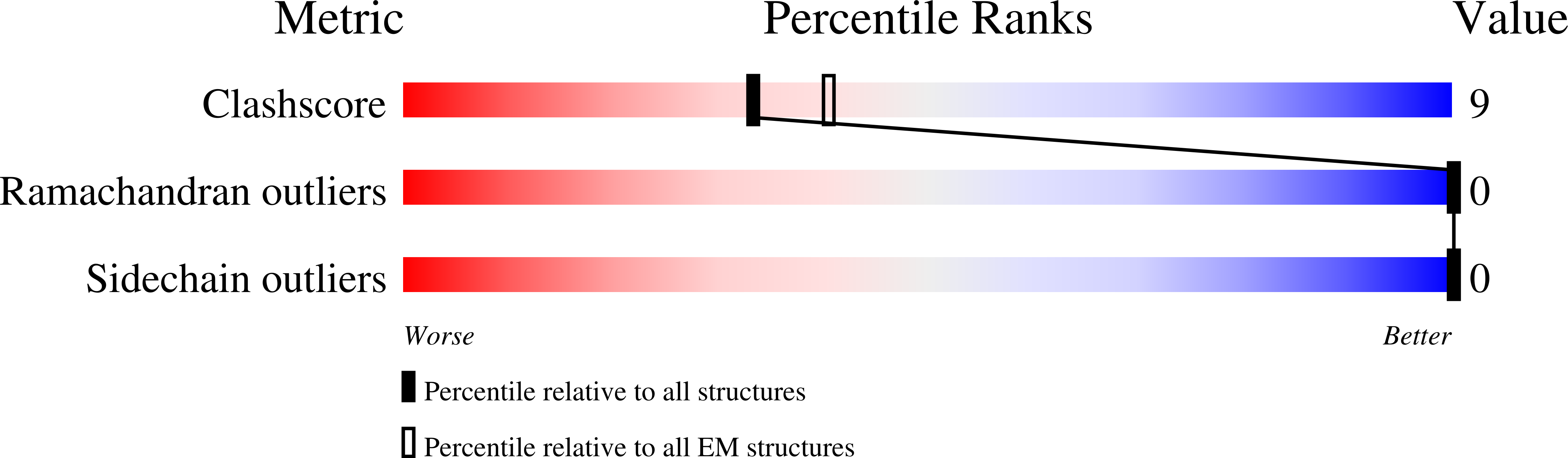Structure and catalytic mechanism of a human triacylglycerol-synthesis enzyme.
Sui, X., Wang, K., Gluchowski, N.L., Elliott, S.D., Liao, M., Walther, T.C., Farese Jr., R.V.(2020) Nature 581: 323-328
- PubMed: 32433611
- DOI: https://doi.org/10.1038/s41586-020-2289-6
- Primary Citation of Related Structures:
6VYI, 6VZ1 - PubMed Abstract:
Triacylglycerols store metabolic energy in organisms and have industrial uses as foods and fuels. Excessive accumulation of triacylglycerols in humans causes obesity and is associated with metabolic diseases 1 . Triacylglycerol synthesis is catalysed by acyl-CoA diacylglycerol acyltransferase (DGAT) enzymes 2-4 , the structures and catalytic mechanisms of which remain unknown. Here we determined the structure of dimeric human DGAT1, a member of the membrane-bound O-acyltransferase (MBOAT) family, by cryo-electron microscopy at approximately 3.0 Å resolution. DGAT1 forms a homodimer through N-terminal segments and a hydrophobic interface, with putative active sites within the membrane region. A structure obtained with oleoyl-CoA substrate resolved at approximately 3.2 Å shows that the CoA moiety binds DGAT1 on the cytosolic side and the acyl group lies deep within a hydrophobic channel, positioning the acyl-CoA thioester bond near an invariant catalytic histidine residue. The reaction centre is located inside a large cavity, which opens laterally to the membrane bilayer, providing lipid access to the active site. A lipid-like density-possibly representing an acyl-acceptor molecule-is located within the reaction centre, orthogonal to acyl-CoA. Insights provided by the DGAT1 structures, together with mutagenesis and functional studies, provide the basis for a model of the catalysis of triacylglycerol synthesis by DGAT.
Organizational Affiliation:
Department of Molecular Metabolism, Harvard T. H. Chan School of Public Health, Boston, MA, USA.