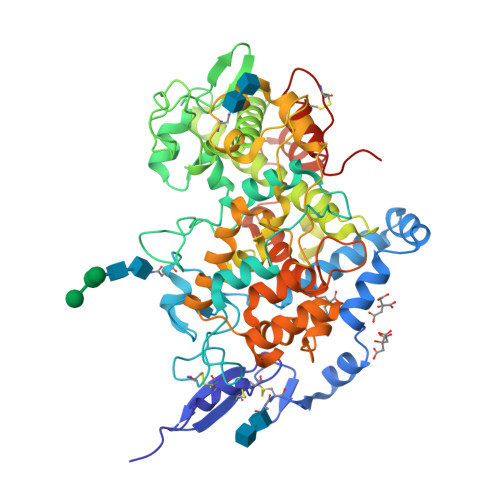
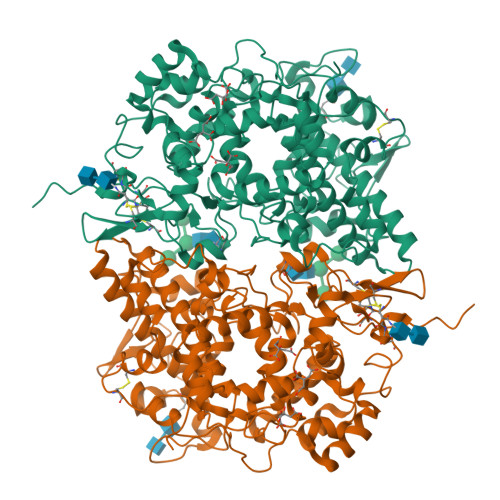
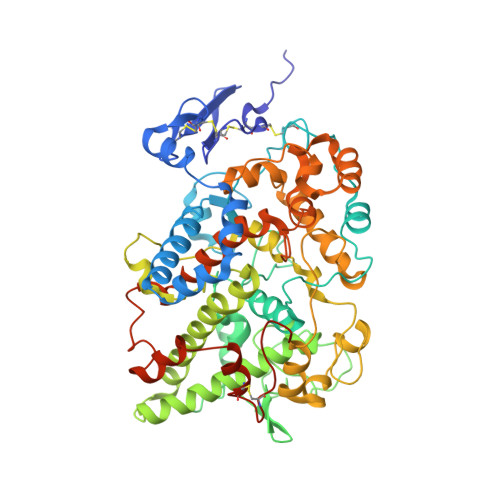
Three-dimensional structure of human cyclooxygenase (hCOX)-1.
Miciaccia, M., Belviso, B.D., Iaselli, M., Cingolani, G., Ferorelli, S., Cappellari, M., Loguercio Polosa, P., Perrone, M.G., Caliandro, R., Scilimati, A.(2021) Sci Rep 11: 4312-4312
- PubMed: 33619313
- DOI: https://doi.org/10.1038/s41598-021-83438-z
- Primary Citation of Related Structures:
6Y3C - PubMed Abstract:
The beneficial effects of Cyclooxygenases (COX) inhibitors on human health have been known for thousands of years. Nevertheless, COXs, particularly COX-1, have been linked to a plethora of human diseases such as cancer, heart failure, neurological and neurodegenerative diseases only recently. COXs catalyze the first step in the biosynthesis of prostaglandins (PGs) and are among the most important mediators of inflammation. All published structural work on COX-1 deals with the ovine isoenzyme, which is easier to produce in milligram-quantities than the human enzyme and crystallizes readily. Here, we report the long-sought structure of the human cyclooxygenase-1 (hCOX-1) that we refined to an R/R free of 20.82/26.37, at 3.36 Å resolution. hCOX-1 structure provides a detailed picture of the enzyme active site and the residues crucial for inhibitor/substrate binding and catalytic activity. We compared hCOX-1 crystal structure with the ovine COX-1 and human COX-2 structures by using metrics based on Cartesian coordinates, backbone dihedral angles, and solvent accessibility coupled with multivariate methods. Differences and similarities among structures are discussed, with emphasis on the motifs responsible for the diversification of the various enzymes (primary structure, stability, catalytic activity, and specificity). The structure of hCOX-1 represents an essential step towards the development of new and more selective COX-1 inhibitors of enhanced therapeutic potential.
Organizational Affiliation:
Department of Pharmacy - Pharmaceutical Sciences, University of Bari "Aldo Moro", Via E. Orabona 4, 70125, Bari, Italy.