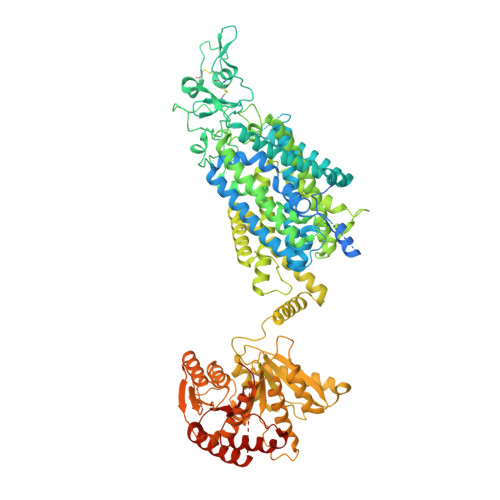
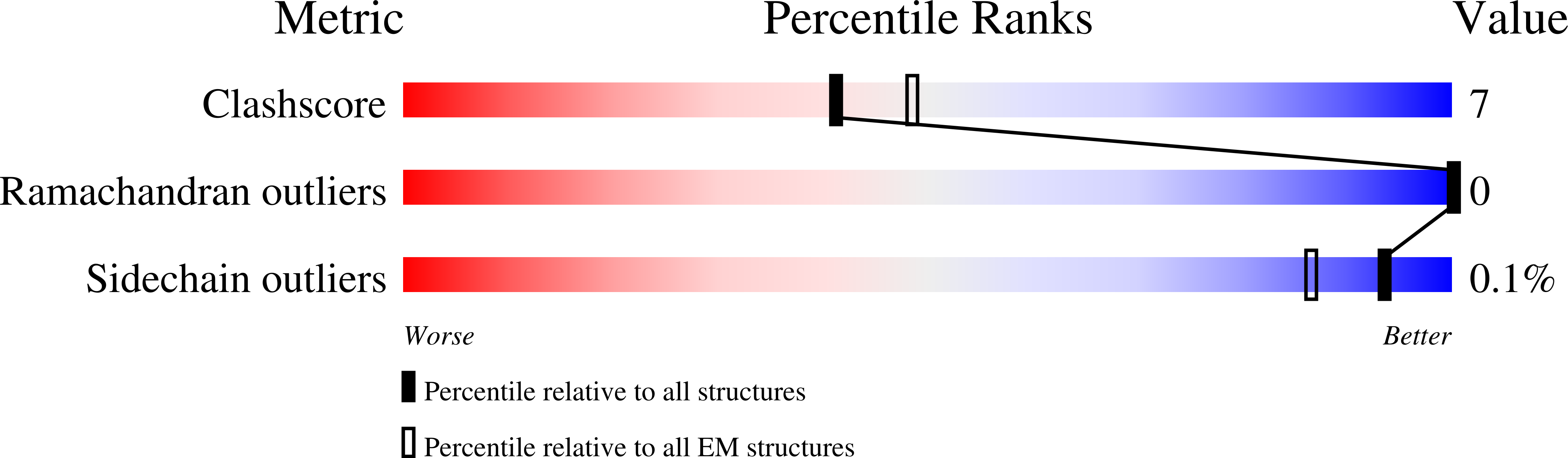Structure of Human Potassium Chloride Transporter KCC3 in NaCl
Chi, G., Man, H., Ebenhoch, R., Reggiano, G., Pike, A.C.W., Wang, D., McKinley, G., Mukhopadhyay, S.M.M., Chalk, R., Moreau, C., Snee, M., Bohstedt, T., Singh, N.K., Abrusci, P., Arrowsmith, C.H., Bountra, C., Edwards, A.M., Marsden, B.D., Burgess-Brown, N.A., DiMaio, F., Duerr, K.L., Structural Genomics Consortium (SGC)To be published.