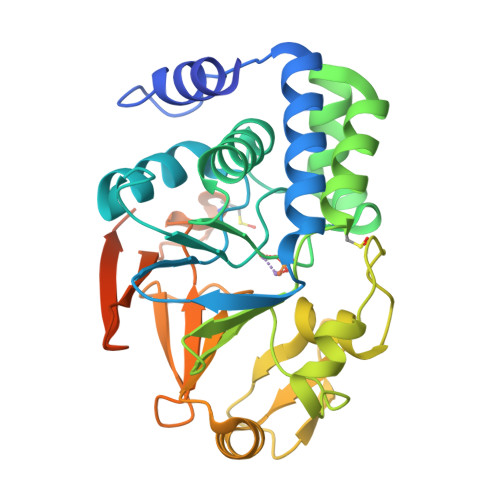
Towards Dissecting the Mechanism of Protein Phosphatase-1 Inhibition by Its C-Terminal Phosphorylation.
Salvi, F., Hoermann, B., Del Pino Garcia, J., Fontanillo, M., Derua, R., Beullens, M., Bollen, M., Barabas, O., Kohn, M.(2021) Chembiochem 22: 834-838
- PubMed: 33085143
- DOI: https://doi.org/10.1002/cbic.202000669
- Primary Citation of Related Structures:
6ZK6 - PubMed Abstract:
Phosphoprotein phosphatase-1 (PP1) is a key player in the regulation of phospho-serine (pSer) and phospho-threonine (pThr) dephosphorylation and is involved in a large fraction of cellular signaling pathways. Aberrant activity of PP1 has been linked to many diseases, including cancer and heart failure. Besides a well-established activity control by regulatory proteins, an inhibitory function for phosphorylation (p) of a Thr residue in the C-terminal intrinsically disordered tail of PP1 has been demonstrated. The associated phenotype of cell-cycle arrest was repeatedly proposed to be due to autoinhibition of PP1 through either conformational changes or substrate competition. Here, we use PP1 variants created by mutations and protein semisynthesis to differentiate between these hypotheses. Our data support the hypothesis that pThr exerts its inhibitory function by mediating protein complex formation rather than by a direct mechanism of structural changes or substrate competition.
Organizational Affiliation:
Genome Biology Unit, European Molecular Biology Laboratory, Meyerhofstraße 1, 69117, Heidelberg, Germany.