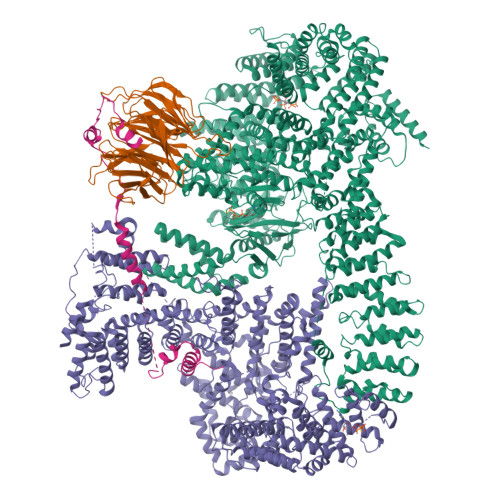
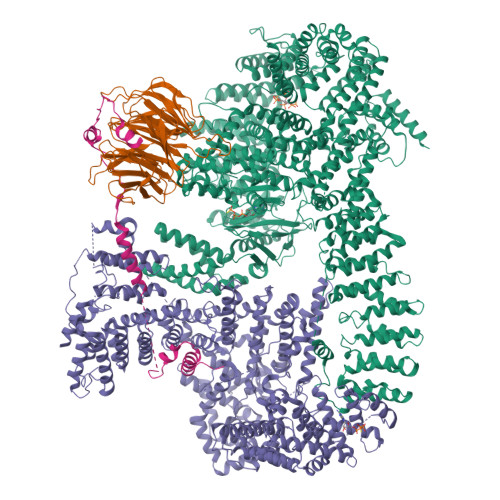
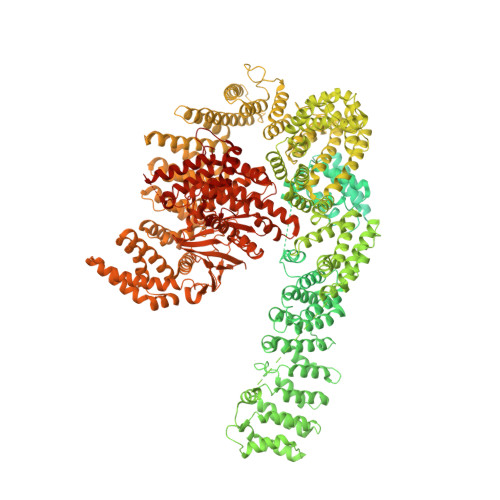
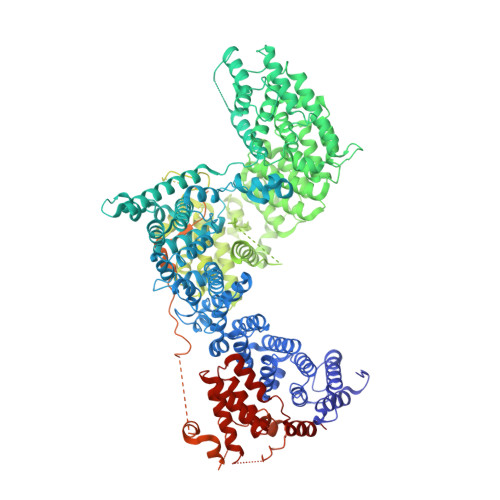
The 3.2- angstrom resolution structure of human mTORC2.
Scaiola, A., Mangia, F., Imseng, S., Boehringer, D., Berneiser, K., Shimobayashi, M., Stuttfeld, E., Hall, M.N., Ban, N., Maier, T.(2020) Sci Adv 6
- PubMed: 33158864
- DOI: https://doi.org/10.1126/sciadv.abc1251
- Primary Citation of Related Structures:
6ZWM, 6ZWO - PubMed Abstract:
The protein kinase mammalian target of rapamycin (mTOR) is the central regulator of cell growth. Aberrant mTOR signaling is linked to cancer, diabetes, and neurological disorders. mTOR exerts its functions in two distinct multiprotein complexes, mTORC1 and mTORC2. Here, we report a 3.2-Å resolution cryo-EM reconstruction of mTORC2. It reveals entangled folds of the defining Rictor and the substrate-binding SIN1 subunits, identifies the carboxyl-terminal domain of Rictor as the source of the rapamycin insensitivity of mTORC2, and resolves mechanisms for mTORC2 regulation by complex destabilization. Two previously uncharacterized small-molecule binding sites are visualized, an inositol hexakisphosphate (InsP6) pocket in mTOR and an mTORC2-specific nucleotide binding site in Rictor, which also forms a zinc finger. Structural and biochemical analyses suggest that InsP6 and nucleotide binding do not control mTORC2 activity directly but rather have roles in folding or ternary interactions. These insights provide a firm basis for studying mTORC2 signaling and for developing mTORC2-specific inhibitors.
Organizational Affiliation:
Institute for Molecular Biology and Biophysics, ETH Zurich, Zurich, Switzerland.