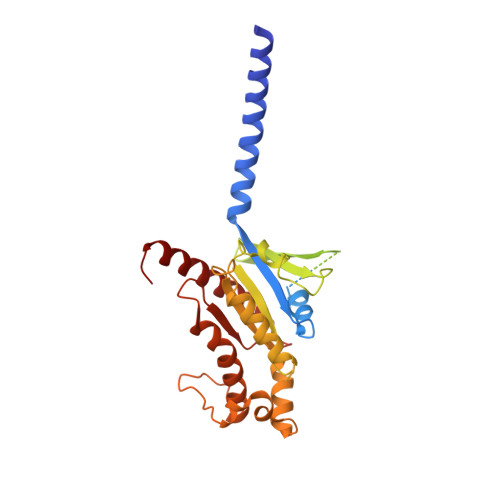
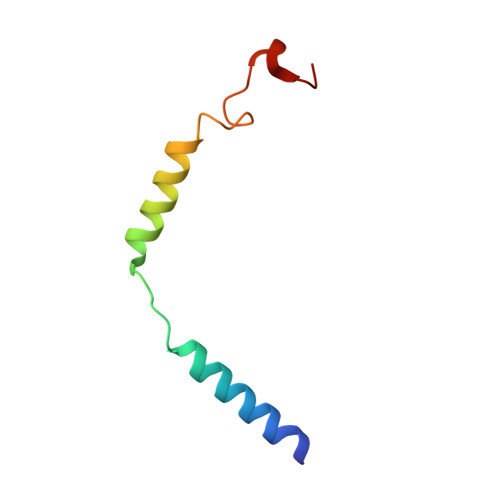
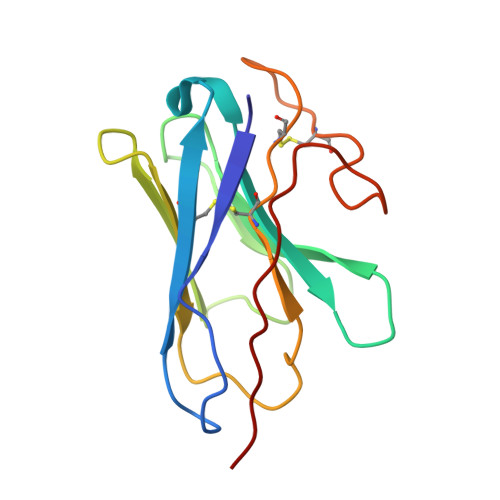
Structural basis of GPBAR activation and bile acid recognition.
Yang, F., Mao, C., Guo, L., Lin, J., Ming, Q., Xiao, P., Wu, X., Shen, Q., Guo, S., Shen, D.D., Lu, R., Zhang, L., Huang, S., Ping, Y., Zhang, C., Ma, C., Zhang, K., Liang, X., Shen, Y., Nan, F., Yi, F., Luca, V.C., Zhou, J., Jiang, C., Sun, J.P., Xie, X., Yu, X., Zhang, Y.(2020) Nature 587: 499-504
- PubMed: 32698187
- DOI: https://doi.org/10.1038/s41586-020-2569-1
- Primary Citation of Related Structures:
7CFM, 7CFN - PubMed Abstract:
The G-protein-coupled bile acid receptor (GPBAR) conveys the cross-membrane signalling of a vast variety of bile acids and is a signalling hub in the liver-bile acid-microbiota-metabolism axis 1-3 . Here we report the cryo-electron microscopy structures of GPBAR-G s complexes stabilized by either the high-affinity P395 4 or the semisynthesized bile acid derivative INT-777 1,3 at 3 Å resolution. These structures revealed a large oval pocket that contains several polar groups positioned to accommodate the amphipathic cholic core of bile acids, a fingerprint of key residues to recognize diverse bile acids in the orthosteric site, a putative second bile acid-binding site with allosteric properties and structural features that contribute to bias properties. Moreover, GPBAR undertakes an atypical mode of activation and G protein coupling that features a different set of key residues connecting the ligand-binding pocket to the G s -coupling site, and a specific interaction motif that is localized in intracellular loop 3. Overall, our study not only reveals unique structural features of GPBAR that are involved in bile acid recognition and allosteric effects, but also suggests the presence of distinct connecting mechanisms between the ligand-binding pocket and the G-protein-binding site in the G-protein-coupled receptor superfamily.
Organizational Affiliation:
Key Laboratory Experimental Teratology of the Ministry of Education, Department of Biochemistry and Molecular Biology, School of Basic Medical Sciences, Cheeloo College of Medicine, Shandong University, Jinan, China.