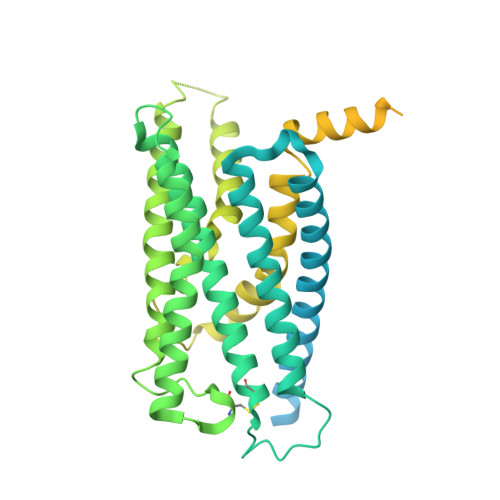
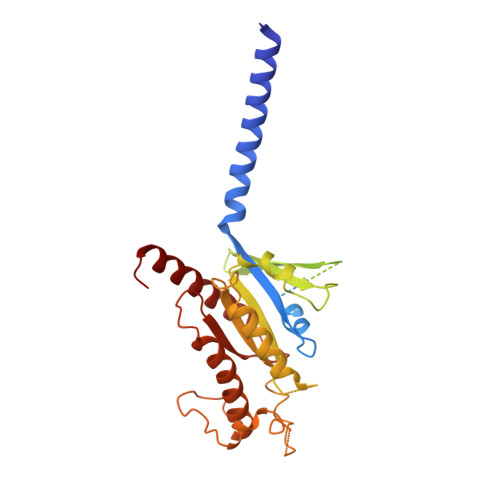
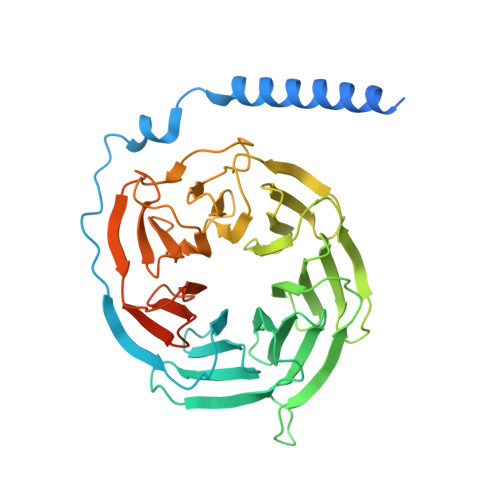
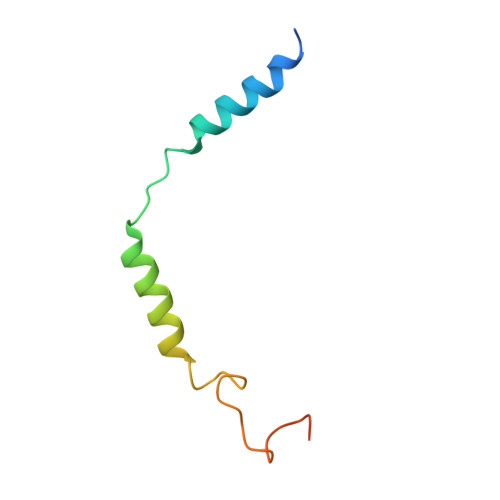
Structural basis for activation of the growth hormone-releasing hormone receptor.
Zhou, F., Zhang, H., Cong, Z., Zhao, L.H., Zhou, Q., Mao, C., Cheng, X., Shen, D.D., Cai, X., Ma, C., Wang, Y., Dai, A., Zhou, Y., Sun, W., Zhao, F., Zhao, S., Jiang, H., Jiang, Y., Yang, D., Eric Xu, H., Zhang, Y., Wang, M.W.(2020) Nat Commun 11: 5205-5205
- PubMed: 33060564
- DOI: https://doi.org/10.1038/s41467-020-18945-0
- Primary Citation of Related Structures:
7CZ5 - PubMed Abstract:
Growth hormone-releasing hormone (GHRH) regulates the secretion of growth hormone that virtually controls metabolism and growth of every tissue through its binding to the cognate receptor (GHRHR). Malfunction in GHRHR signaling is associated with abnormal growth, making GHRHR an attractive therapeutic target against dwarfism (e.g., isolated growth hormone deficiency, IGHD), gigantism, lipodystrophy and certain cancers. Here, we report the cryo-electron microscopy (cryo-EM) structure of the human GHRHR bound to its endogenous ligand and the stimulatory G protein at 2.6 Å. This high-resolution structure reveals a characteristic hormone recognition pattern of GHRH by GHRHR, where the α-helical GHRH forms an extensive and continuous network of interactions involving all the extracellular loops (ECLs), all the transmembrane (TM) helices except TM4, and the extracellular domain (ECD) of GHRHR, especially the N-terminus of GHRH that engages a broad set of specific interactions with the receptor. Mutagenesis and molecular dynamics (MD) simulations uncover detailed mechanisms by which IGHD-causing mutations lead to the impairment of GHRHR function. Our findings provide insights into the molecular basis of peptide recognition and receptor activation, thereby facilitating the development of structure-based drug discovery and precision medicine.
Organizational Affiliation:
The CAS Key Laboratory of Receptor Research, Shanghai Institute of Materia Medica, Chinese Academy of Sciences, 201203, Shanghai, China.