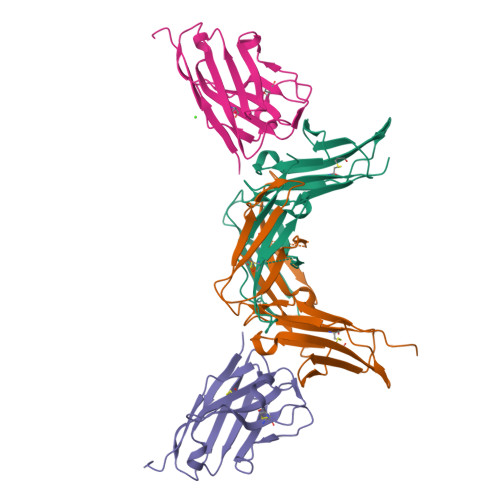
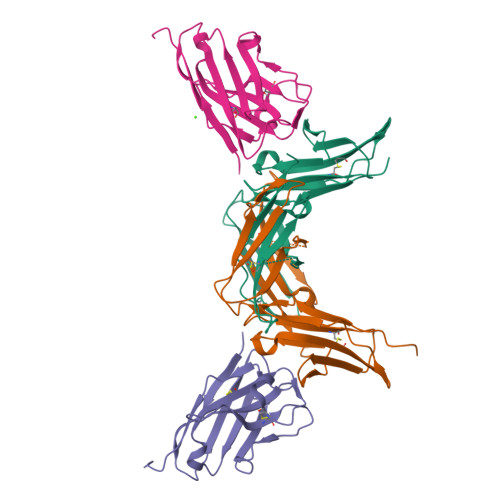
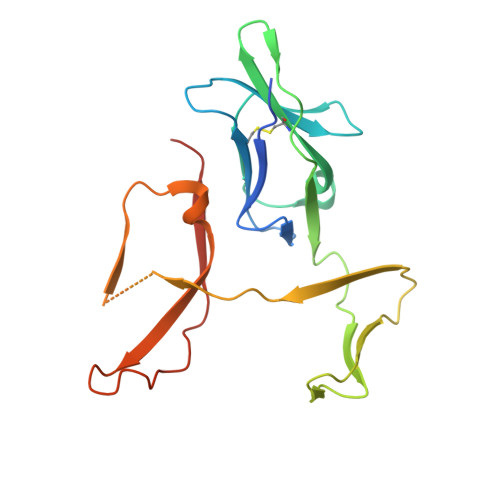
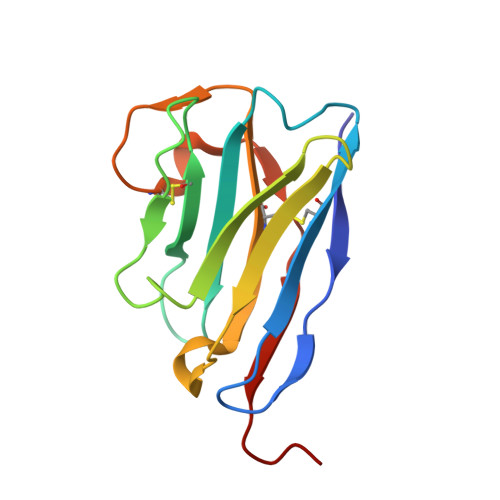
Structural characterization of a novel GPVI-nanobody complex reveals a biologically active domain-swapped GPVI dimer.
Slater, A., Di, Y., Clark, J.C., Jooss, N.J., Martin, E.M., Alenazy, F., Thomas, M.R., Ariens, R.A.S., Herr, A.B., Poulter, N.S., Emsley, J., Watson, S.P.(2021) Blood 137: 3443-3453
- PubMed: 33512486
- DOI: https://doi.org/10.1182/blood.2020009440
- Primary Citation of Related Structures:
7NMU - PubMed Abstract:
Glycoprotein VI (GPVI) is the major signaling receptor for collagen on platelets. We have raised 54 nanobodies (Nb), grouped into 33 structural classes based on their complementary determining region 3 loops, against recombinant GPVI-Fc (dimeric GPVI) and have characterized their ability to bind recombinant GPVI, resting and activated platelets, and to inhibit platelet activation by collagen. Nbs from 6 different binding classes showed the strongest binding to recombinant GPVI-Fc, suggesting that there was not a single dominant class. The most potent 3, Nb2, 21, and 35, inhibited collagen-induced platelet aggregation with nanomolar half maximal inhibitory concentration (IC50) values and inhibited platelet aggregation under flow. The binding KD of the most potent Nb, Nb2, against recombinant monomeric and dimeric GPVI was 0.6 and 0.7 nM, respectively. The crystal structure of monomeric GPVI in complex with Nb2 revealed a binding epitope adjacent to the collagen-related peptide (CRP) binding groove within the D1 domain. In addition, a novel conformation of GPVI involving a domain swap between the D2 domains was observed. The domain swap is facilitated by the outward extension of the C-C' loop, which forms the domain swap hinge. The functional significance of this conformation was tested by truncating the hinge region so that the domain swap cannot occur. Nb2 was still able to displace collagen and CRP binding to the mutant, but signaling was abolished in a cell-based NFAT reporter assay. This demonstrates that the C-C' loop region is important for GPVI signaling but not ligand binding and suggests the domain-swapped structure may represent an active GPVI conformation.
Organizational Affiliation:
Institute of Cardiovascular Sciences, College of Medical and Dental Sciences, University of Birmingham, Birmingham, United Kingdom.