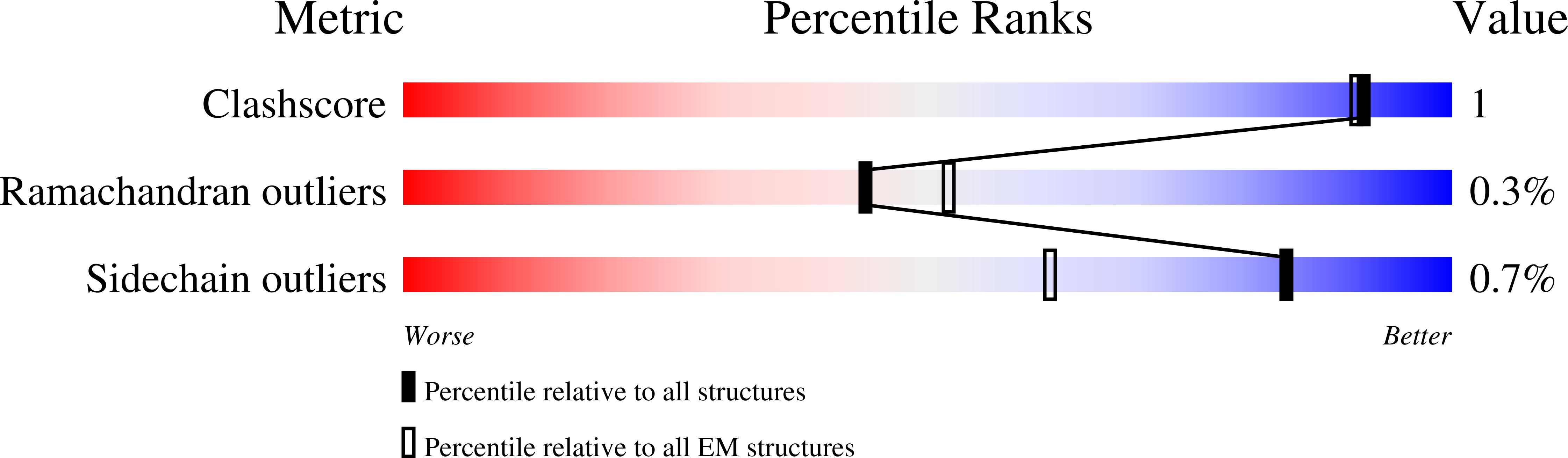Streptomyces umbrella toxin particles block hyphal growth of competing species.
Zhao, Q., Bertolli, S., Park, Y.J., Tan, Y., Cutler, K.J., Srinivas, P., Asfahl, K.L., Fonesca-Garcia, C., Gallagher, L.A., Li, Y., Wang, Y., Coleman-Derr, D., DiMaio, F., Zhang, D., Peterson, S.B., Veesler, D., Mougous, J.D.(2024) Nature
- PubMed: 38632398
- DOI: https://doi.org/10.1038/s41586-024-07298-z
- Primary Citation of Related Structures:
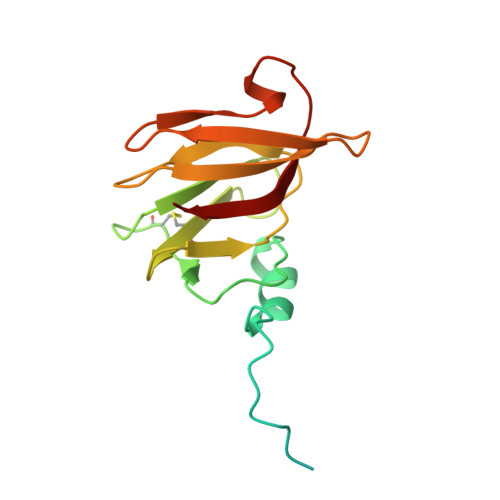
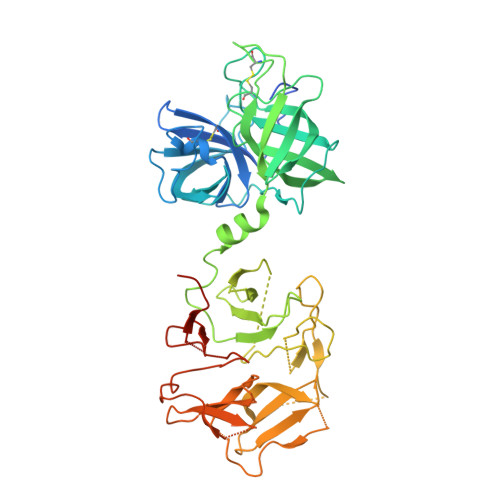
8W20, 8W22 - PubMed Abstract:
Streptomyces are a genus of ubiquitous soil bacteria from which the majority of clinically utilized antibiotics derive 1 . The production of these antibacterial molecules reflects the relentless competition Streptomyces engage in with other bacteria, including other Streptomyces species 1,2 . Here we show that in addition to small-molecule antibiotics, Streptomyces produce and secrete antibacterial protein complexes that feature a large, degenerate repeat-containing polymorphic toxin protein. A cryo-electron microscopy structure of these particles reveals an extended stalk topped by a ringed crown comprising the toxin repeats scaffolding five lectin-tipped spokes, which led us to name them umbrella particles. Streptomyces coelicolor encodes three umbrella particles with distinct toxin and lectin composition. Notably, supernatant containing these toxins specifically and potently inhibits the growth of select Streptomyces species from among a diverse collection of bacteria screened. For one target, Streptomyces griseus, inhibition relies on a single toxin and that intoxication manifests as rapid cessation of vegetative hyphal growth. Our data show that Streptomyces umbrella particles mediate competition among vegetative mycelia of related species, a function distinct from small-molecule antibiotics, which are produced at the onset of reproductive growth and act broadly 3,4 . Sequence analyses suggest that this role of umbrella particles extends beyond Streptomyces, as we identified umbrella loci in nearly 1,000 species across Actinobacteria.
Organizational Affiliation:
Department of Microbiology, University of Washington, Seattle, WA, USA.