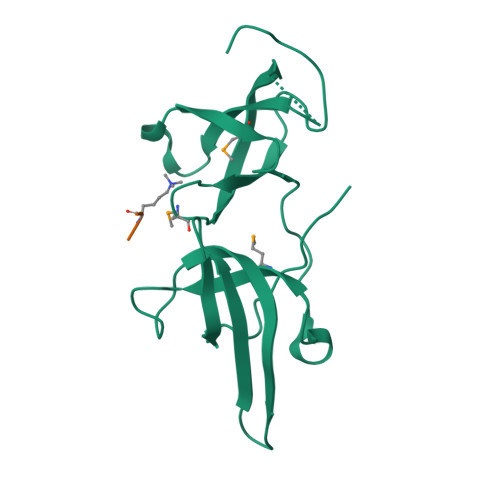
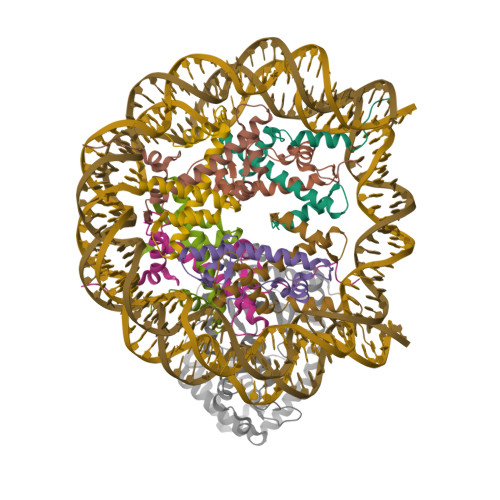
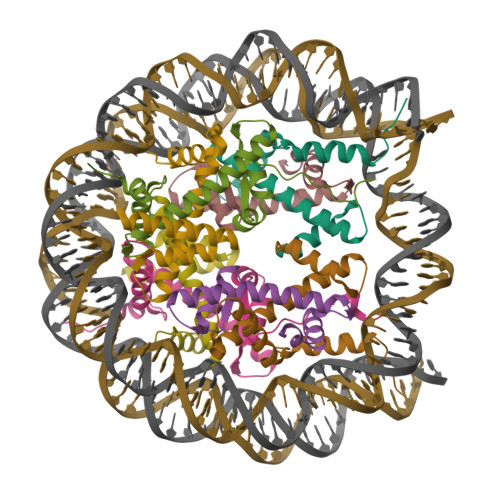
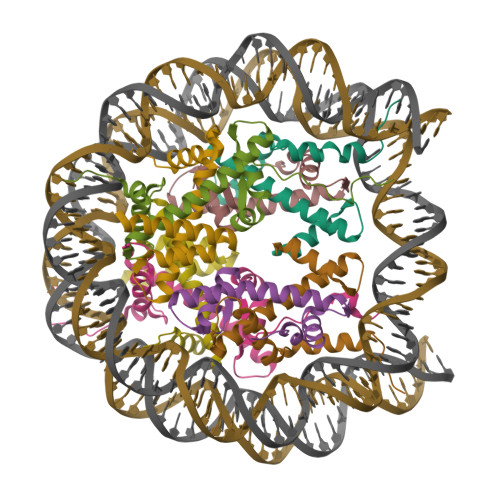
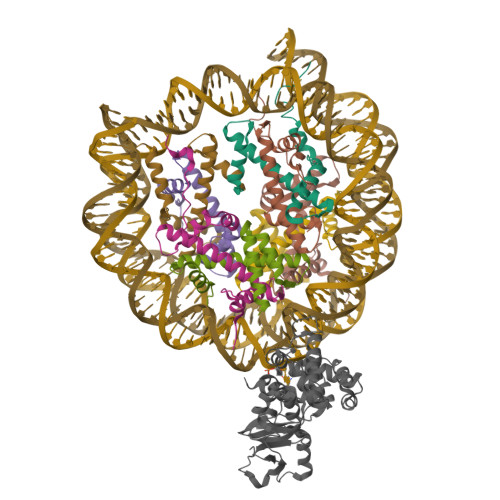
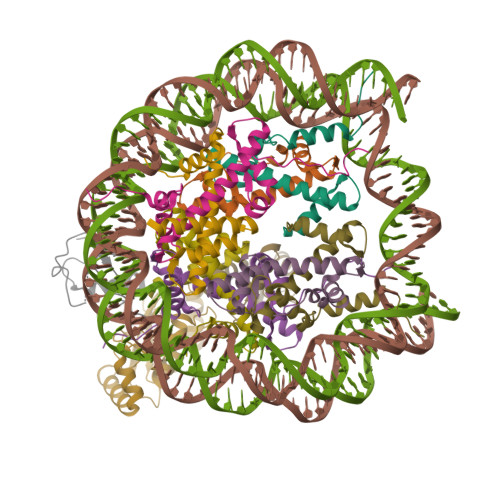
Structure Determination MethodologyScientific Name of Source OrganismRefinement Resolution (Å)Enzyme Classification Name | Walker, J.R., Avvakumov, G.V., Xue, S., Dong, A., Li, Y., Bountra, C., Weigelt, J., Arrowsmith, C.H., Edwards, A.M., Bochkarev, A., Dhe-Paganon, S., Structural Genomics Consortium (SGC) (2011) J Biological Chem 286: 24300-24311 | Released | 2008-09-16 | | Method | X-RAY DIFFRACTION 2.4 Å | | Organisms | | | Macromolecule | |
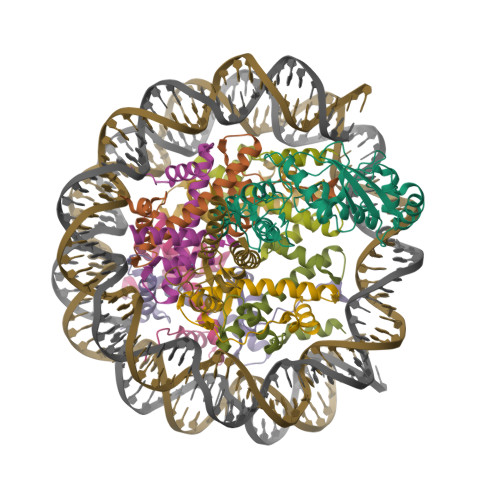
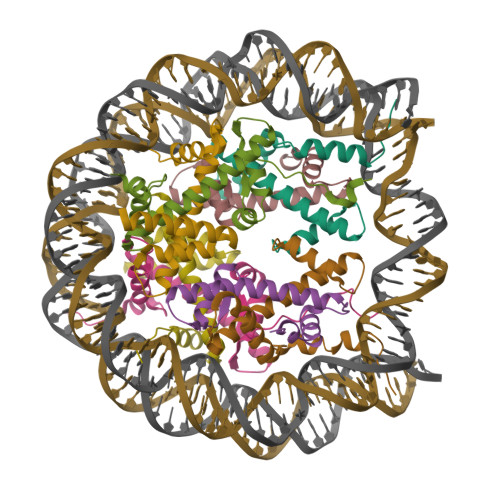
Li, J., Li, Z., Ruan, J., Xu, C., Tong, Y., Pan, P.W., Tempel, W., Crombet, L., Min, J., Zang, J., Structural Genomics Consortium (SGC) (2011) PLoS One 6: e25104-e25104 | Released | 2011-04-06 | | Method | X-RAY DIFFRACTION 2.057 Å | | Organisms | | | Macromolecule | | | Unique Ligands | UNX |
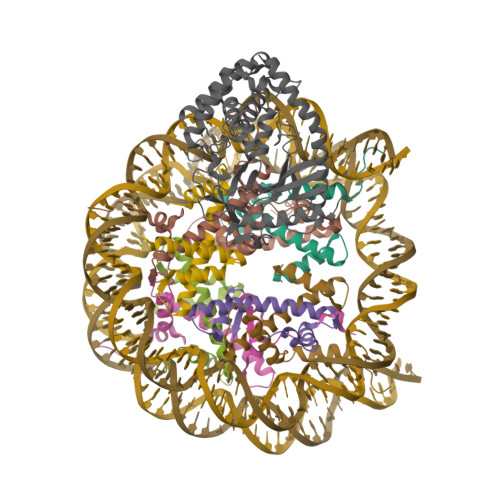
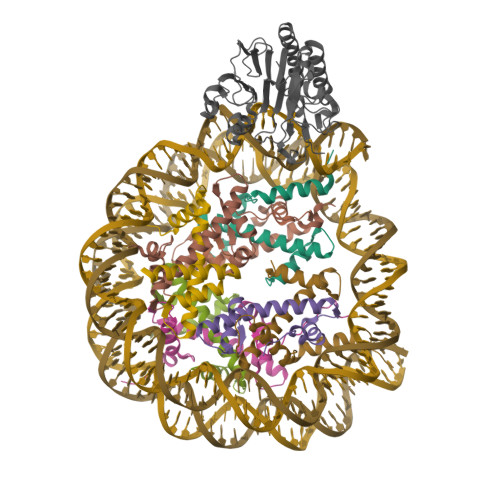
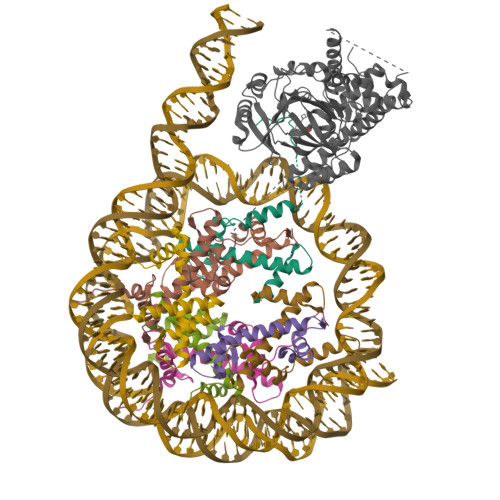
Pengbiao, X., Pingwei, L., Baoyu, Z. (2020) Nature 587: 673-677 | Released | 2020-09-16 | | Method | ELECTRON MICROSCOPY 2.98 Å | | Organisms | | | Macromolecule | Unique protein chains: 5 Unique nucleic acid chains: 2 | | Unique Ligands | ZN |
Xu, P., Li, P., Zhao, B. (2020) Nature 587: 673-677 | Released | 2020-09-16 | | Method | ELECTRON MICROSCOPY 6.8 Å | | Organisms | | | Macromolecule | Unique protein chains: 5 Unique nucleic acid chains: 2 | | Unique Ligands | ZN |
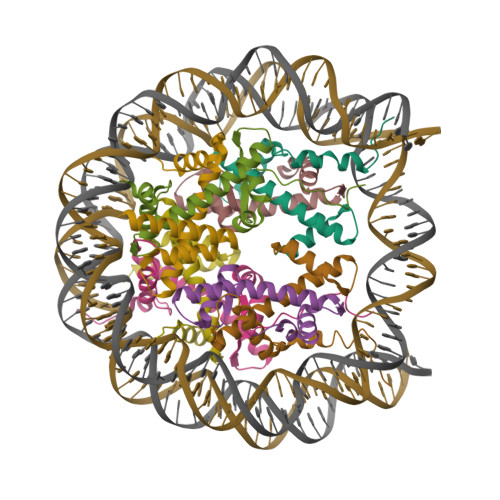
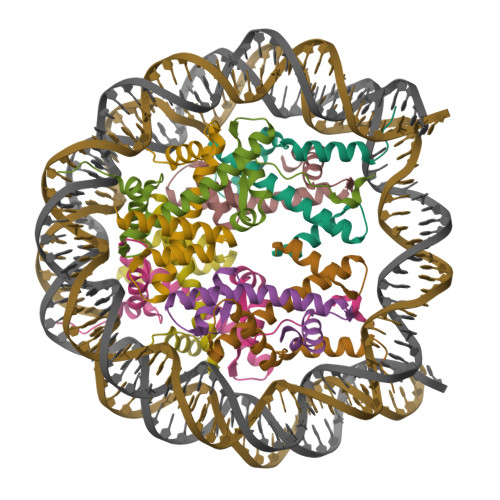
Boyer, J.A., Spangler, C.J., Strauss, J.D., Cesmat, A.P., Liu, P., McGinty, R.K., Zhang, Q. (2020) Science 370: 450-454 | Released | 2020-09-16 | | Method | ELECTRON MICROSCOPY 3.3 Å | | Organisms | | | Macromolecule | Unique protein chains: 5 Unique nucleic acid chains: 2 | | Unique Ligands | ZN |
Boyer, J.A., Spangler, C.J., Strauss, J.D., Cesmat, A.P., Liu, P., McGinty, R.K., Zhang, Q. (2020) Science 370: 450-454 | Released | 2020-09-16 | | Method | ELECTRON MICROSCOPY 3.3 Å | | Organisms | | | Macromolecule | Unique protein chains: 5 Unique nucleic acid chains: 2 | | Unique Ligands | ZN |
Witus, S.R., Burrell, A.L., Hansen, J.M., Farrell, D.P., Dimaio, F., Kollman, J.M., Klevit, R.E. (2021) Nat Struct Mol Biol 28: 268-277 | Released | 2021-02-17 | | Method | ELECTRON MICROSCOPY 3.9 Å | | Organisms | | | Macromolecule | Unique protein chains: 6 Unique nucleic acid chains: 2 | | Unique Ligands | ZN |
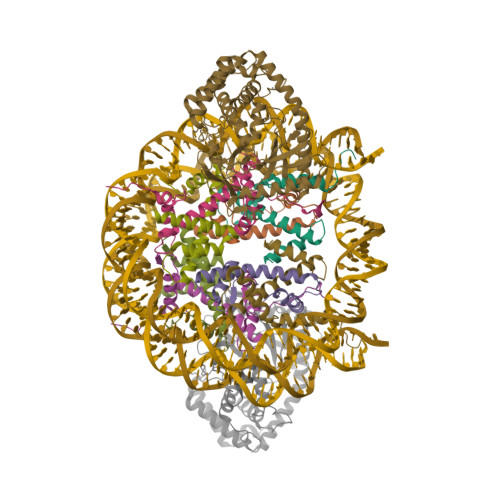
Weaver, T.M., Freudenthal, B.D. (2022) Nat Commun 13: 5390-5390 | Released | 2022-09-07 | | Method | ELECTRON MICROSCOPY 3.4 Å | | Organisms | | | Macromolecule | Unique protein chains: 5 Unique nucleic acid chains: 2 |
Freudenthal, B.D., Weaver, T.M. (2022) Nat Commun 13: 5390-5390 | Released | 2022-09-07 | | Method | ELECTRON MICROSCOPY 3.1 Å | | Organisms | | | Macromolecule | Unique protein chains: 4 Unique nucleic acid chains: 2 |
Freudenthal, B.D., Weaver, T.M. (2022) Nat Commun 13: 5390-5390 | Released | 2022-09-07 | | Method | ELECTRON MICROSCOPY 3.4 Å | | Organisms | | | Macromolecule | Unique protein chains: 4 Unique nucleic acid chains: 2 |
Freudenthal, B.D., Weaver, T.M. (2022) Nat Commun 13: 5390-5390 | Released | 2022-09-07 | | Method | ELECTRON MICROSCOPY 4 Å | | Organisms | | | Macromolecule | Unique protein chains: 4 Unique nucleic acid chains: 2 |
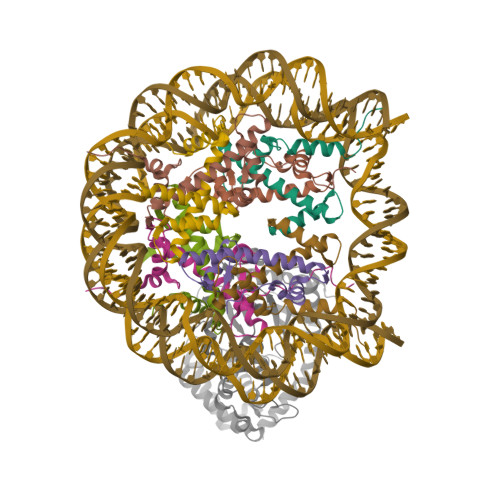
Spangler, C.J., Skrajna, A., Foley, C.A., Budziszewski, G.R., Azzam, D.N., James, L.I., Frye, S.V., McGinty, R.K. (2023) Nat Chem Biol 19: 624-632 | Released | 2023-02-22 | | Method | ELECTRON MICROSCOPY 3.2 Å | | Organisms | | | Macromolecule | Unique protein chains: 5 Unique nucleic acid chains: 2 | | Unique Ligands | FE, OH0 |
Oishi, T., Hatazawa, S., Kujirai, T., Kato, J., Kobayashi, Y., Ogasawara, M., Akatsu, M., Takizawa, Y., Kurumizaka, H. (2023) Nucleic Acids Res 51: 10364-10374 | Released | 2023-10-04 | | Method | ELECTRON MICROSCOPY 2.36 Å | | Organisms | | | Macromolecule | Unique protein chains: 4 Unique nucleic acid chains: 2 |
Oishi, T., Hatazawa, S., Kujirai, T., Kato, J., Kobayashi, Y., Ogasawara, M., Akatsu, M., Takizawa, Y., Kurumizaka, H. (2023) Nucleic Acids Res 51: 10364-10374 | Released | 2023-10-04 | | Method | ELECTRON MICROSCOPY 2.48 Å | | Organisms | | | Macromolecule | Unique protein chains: 4 Unique nucleic acid chains: 2 |
Xu, P.B., Ablasser, A. (2024) Nature 627: 873-879 | Released | 2024-02-14 | | Method | ELECTRON MICROSCOPY 3.5 Å | | Organisms | | | Macromolecule | Unique protein chains: 10 Unique nucleic acid chains: 2 | | Unique Ligands | ZN |
Cookis, T., Nogales, E. (2025) Nat Struct Mol Biol 32: 393-404 | Released | 2025-01-15 | | Method | ELECTRON MICROSCOPY 3.1 Å | | Organisms | | | Macromolecule | Unique protein chains: 4 Unique nucleic acid chains: 2 |
Cookis, T., Nogales, E. (2025) Nat Struct Mol Biol 32: 393-404 | Released | 2025-01-22 | | Method | ELECTRON MICROSCOPY 3.5 Å | | Organisms | | | Macromolecule | Unique protein chains: 5 Unique nucleic acid chains: 2 |
Cookis, T., Nogales, E. (2025) Nat Struct Mol Biol 32: 393-404 | Released | 2025-01-22 | | Method | ELECTRON MICROSCOPY 3.3 Å | | Organisms | | | Macromolecule | Unique protein chains: 6 Unique nucleic acid chains: 2 |
Cookis, T., Nogales, E. (2025) Nat Struct Mol Biol 32: 393-404 | Released | 2025-01-15 | | Method | ELECTRON MICROSCOPY 3.1 Å | | Organisms | | | Macromolecule | Unique protein chains: 4 Unique nucleic acid chains: 2 |
Weaver, T.M., Ling, J.A., Freudenthal, B.D. (2024) Nat Commun 15: 10722-10722 | Released | 2024-10-30 | | Method | ELECTRON MICROSCOPY 3.1 Å | | Organisms | | | Macromolecule | Unique protein chains: 4 Unique nucleic acid chains: 2 |
Weaver, T.M., Ling, J.A., Freudenthal, B.D. (2024) Nat Commun 15: 10722-10722 | Released | 2024-10-30 | | Method | ELECTRON MICROSCOPY 3.3 Å | | Organisms | | | Macromolecule | Unique protein chains: 5 Unique nucleic acid chains: 2 |
Weaver, T.M., Ling, J.A., Freudenthal, B.D. (2024) Nat Commun 15: 10722-10722 | Released | 2024-10-30 | | Method | ELECTRON MICROSCOPY 3 Å | | Organisms | | | Macromolecule | Unique protein chains: 4 Unique nucleic acid chains: 2 |
Weaver, T.M., Ling, J.A., Freudenthal, B.D. (2024) Nat Commun 15: 10722-10722 | Released | 2024-10-30 | | Method | ELECTRON MICROSCOPY 3.6 Å | | Organisms | | | Macromolecule | Unique protein chains: 5 Unique nucleic acid chains: 2 |
|