RCSB PDB Help
General Help
Symmetry Resources in the PDB
Introduction
In nature and in structures deposited in the PDB, a majority of proteins interact with each other to form assemblies. These assemblies play many structural and/or functional roles in biology:
- Cooperativity: This phenomenon is seen in oligomers where the binding of a ligand, cofactor, or other molecule causes a change not only at the site of interaction but also in neighboring molecules. For example, the hemoglobin heterotetramer shows cooperativity in binding oxygen. After one of the four hemoglobin subunits binds oxygen, it becomes progressively easier for each of the other subunits in the protein to bind to oxygen. You can read a review (Changeux 2013) for more details about cooperativity.
- Multivalent interactions: Assemblies with multiple binding sites allow multivalent binding to a target. For example, antibodies contain two or more identical binding sites for an antigen, each created by different copies of the proteins in the assembly. These binding sites can strengthen the interaction of an antibody with its target, such as a virus, by binding to several neighboring sites on the target’s surface.
- Building functional shapes: Assemblies are widely used in biology for building large assemblies with specific functional shapes. For example, icosahedral viral capsids are built from many copies of one or a few proteins, and channels through membranes are often built from multiple copies of proteins forming a tube-like shape.
Symmetry in Protein Assemblies
Most often, protein assemblies are symmetrical: they are built of many identical subunits that each interact with their neighbors in identical ways. Symmetrical assemblies have a major advantage over asymmetric assemblies: they are economical to encode in the genome, so many copies of a small protein may be used to build a large assembly. For example, viral capsids are built from many identical proteins (or subunits) arranged with icosahedral symmetry.
Symmetry has fascinated scientists, mathematicians, and artists for centuries, and there is a comprehensive formalism for defining symmetry in an object. In general, symmetrical objects are defined by “symmetry operations.” The simplest symmetry operation is a rotation. For a demonstration, take a piece of blank paper and put it on a table. Then, rotate it by 180 degrees around its center. It will look the same as when you started, so we say that the piece of paper has rotational symmetry. Similarly, other symmetry operations - translation, reflection, reversal, and inversion - describe other ways in which objects can have self-similarity. In biology, since most biomolecules have an intrinsic handedness, only two types of symmetry operators are relevant: rotation and translation. However, combinations of rotation and translation operations still lead to a wide variety of different types of symmetrical biomolecular assemblies.
Symmetry Operations
Oligomeric assemblies in PDB have either point group symmetry or helical symmetry (Goodsell and Olson, 2000). Point group symmetries are defined by one or more axes of rotation that form defined, self-consistent assemblies, while helical symmetries combine rotation and translation to form long, helical fibers. Several types of these symmetries are commonly found in biomolecules:
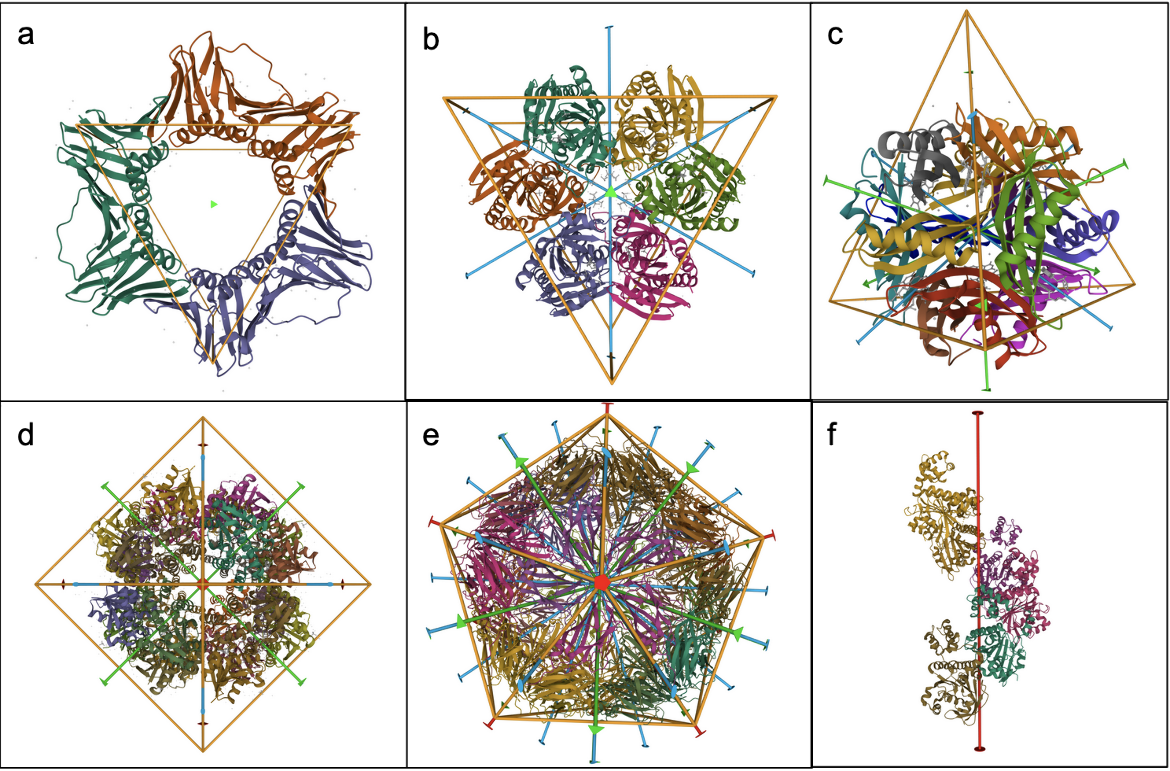
- Cyclic (Cn) symmetry involves the symmetrical placement of n identical molecules or groups of molecules around a single axis of rotation. For example, in C3 symmetry there are 3 molecules that are positioned at 360/n (120) degree intervals around the axis of rotation (Figure 1a).
- Dihedral (Dn) symmetry combines an n-fold cyclic axis of symmetry with n perpendicular 2-fold rotation axes. For example, objects with D3 symmetry have a central 3-fold axis and three perpendicular 2-fold axes, together creating an assembly with 6 identical subunits (Figure 1b).
- Cubic symmetries contain 3-fold rotational axes combined with other, non-perpendicular rotational axes. Three Cubic symmetries are commonly seen in protein assemblies:
- Tetrahedral (T) symmetry has 2-fold axes intersecting the 3-fold axes (Figure 1c)
- Octahedral (O) symmetry has 4-fold axes intersecting the 3-fold axes (Figure 1d)
- Icosahedral (I) symmetry has 5-fold and 2-fold axes intersecting the 3-fold axes (Figure 1e)
- Helical (H) symmetries combine rotational symmetry with translation along the rotation axis (Figure 1f).
Note that in protein crystals, the symmetry of the assembly (point group symmetry or helical symmetry) is combined with additional translational symmetries based on the crystal lattice. Therefore in some of these structures (determined by X-ray crystallography), this can impact the way that coordinates are represented in the PDB archive - see "Introduction to Biological Assemblies" for more information.
Levels of Symmetry
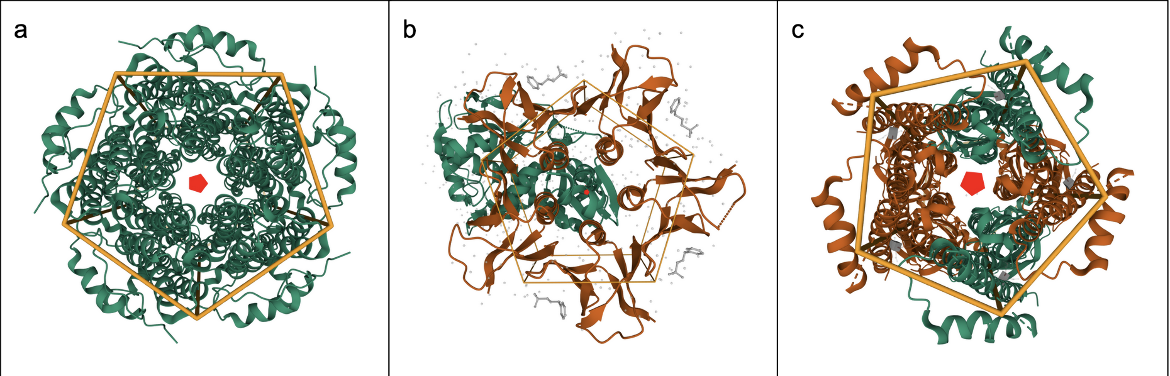
The relationships between molecules in an assembly may either be global (involving the entire structure, Figure 2a), local (involving part of the structure, Figure 2b), or pseudo symmetric (involving apparent but not exact symmetry, Figure 2c). See examples below:
Why explore protein symmetry?
Protein symmetry may influence how the molecules in an assembly facilitate its function. Since symmetric assemblies are stabilized by repeating sets of specific intermolecular interactions, exploring these interactions can provide insights into how an assembly is formed and how its formation may be altered, in turn, allowing regulation of the molecule’s biological function(s).
Documentation: Accessing Protein Symmetry Information
There are several options for exploring protein symmetry in PDB assemblies. Three of them are listed below:
Protein Symmetry Browser
Using the Protein Symmetry Browser, one can view and select from the various symmetry operations and level of symmetry. For details on how to use this browser click here.
Protein Symmetry Annotations
On the Structure Summary Page for every PDB entry, symmetry information for each assembly of the PDB structure is displayed below the image of the assembly. The few types of symmetry annotations for PDB structures are listed below:
- If there is no symmetry in the structure, it is labeled as “Global Symmetry: Asymmetric - C1”.
- “Stoichiometry” indicates the number of proteins participating in the assembly, and whether the assembly comprises multiple copies of the same protein (homomer) or two or more different proteins (heteromer).
- If there is global symmetry, local symmetry, or pseudosymmetry, it is listed here along with a link to a visualization of the symmetry axis or axes.
- The “Find Similar Assemblies” link launches a structure similarity search for the assembly using a strict shape match. The search will identify other structures in the PDB that may have the same or similar symmetry and assembly structure.
Visualizing Protein Symmetry in 3D (using Mol*)
On the Mol* page or (3D View tab of the Structure Summary page) for any structure in the PDB, there is an “Assembly Symmetry” button in the Control (right hand) panel. Clicking on the “Enable” button that appears when the Control panel is selected colors the symmetrically arranged proteins in the same color and displays the symmetry operators for the assembly on the 3D canvas. To visualize the locations of each of the symmetrically positioned proteins and how they interact with each other, the polymers in the assembly may be colored in different colors (e.g., color by chain property >> Chain instance or Entity ID). These coloring options have been used for Figures 1 and 2.
Examples
A Case with Global Symmetry
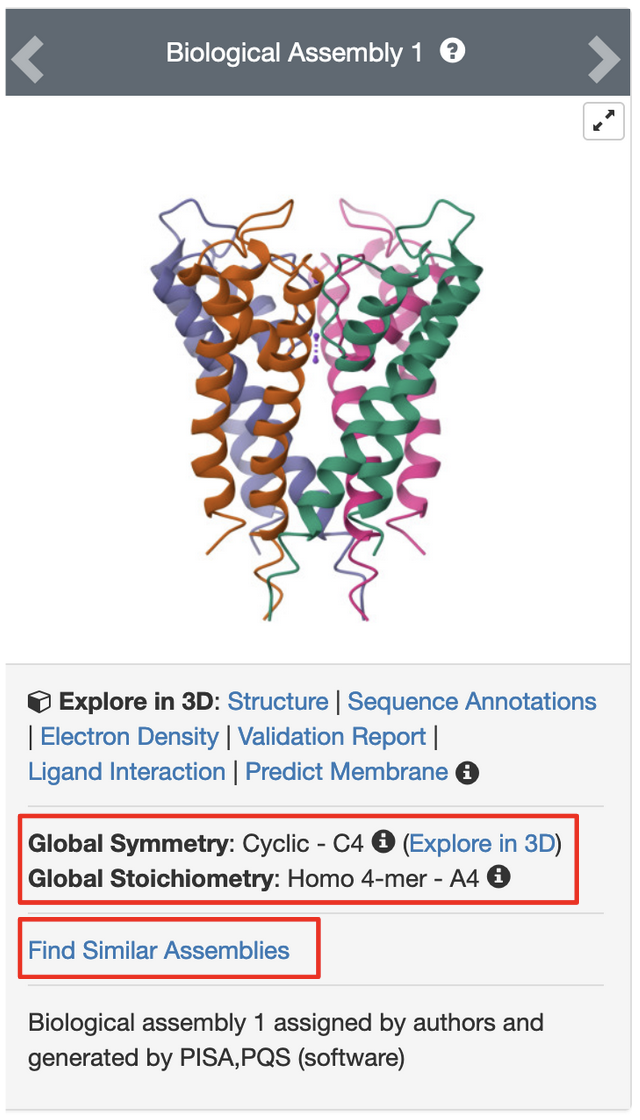
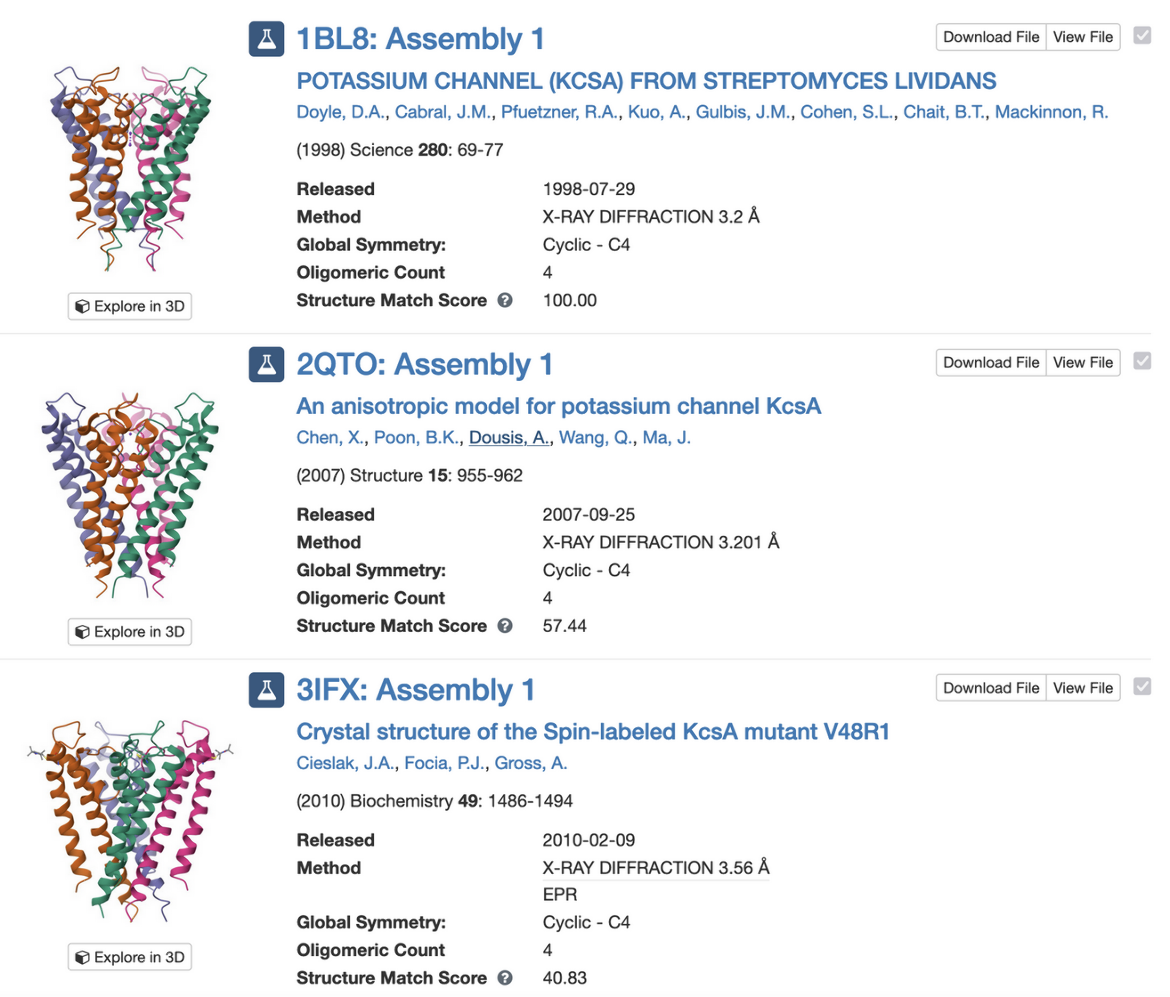
PDB entry 1bl8 is the structure of the tetrameric potassium channel. The symmetry information displayed on the Structure Summary Page is shown here:
Note that the structure in this entry is a homotetramer with Cyclic - C4 symmetry.
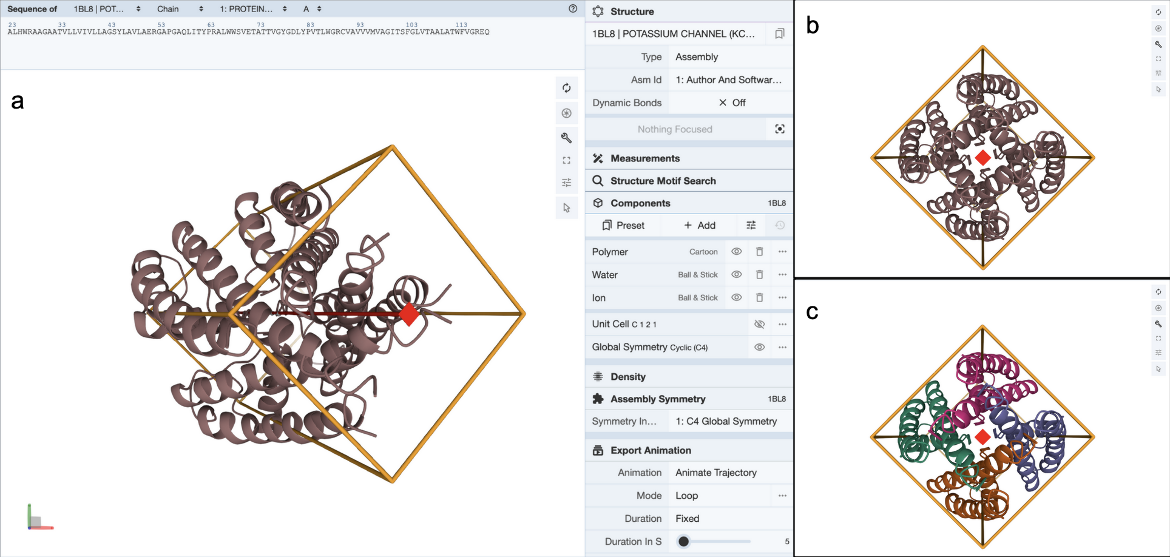
Clicking on the (3D View) link next to the “Global Symmetry: Cyclic - C4” label opens a 3D visualization of the structure in Mol* (Figure 4a). You can rotate the molecule to view down the 4-fold rotation axis (Figure 4b) and color the proteins by chain instance (Figure 4c) to see how the four copies of the proteins are assembled to form the channel.
Clicking on “Find Similar Assemblies” on the Structure Summary Page (Figure 3) launches a strict-match assembly search to find similar assemblies in the PDB (Figure 5).
A Case with Pseudo Symmetry
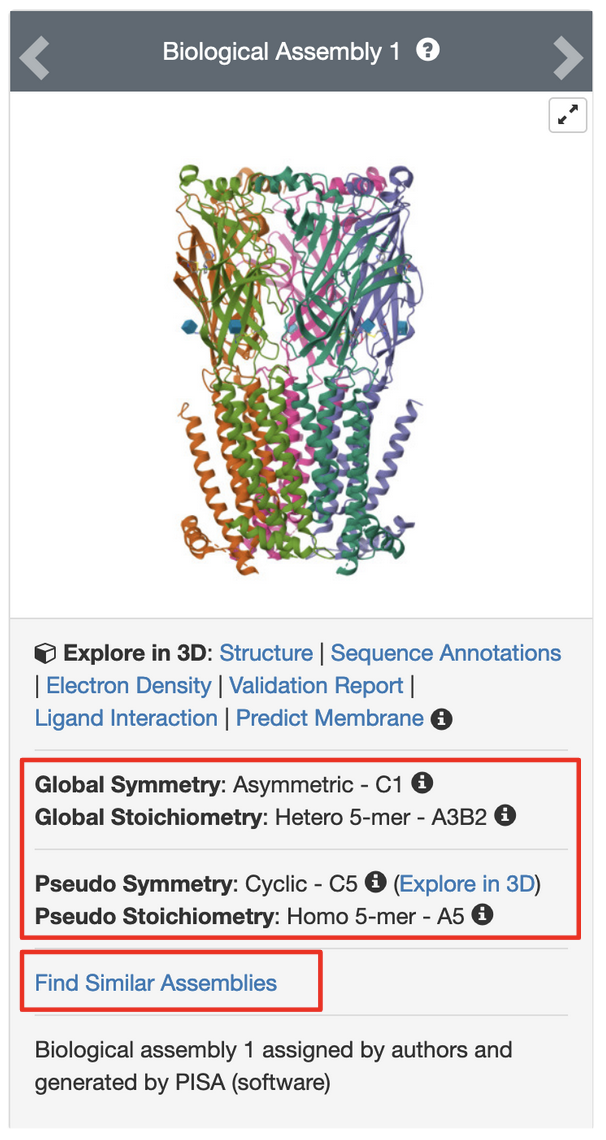
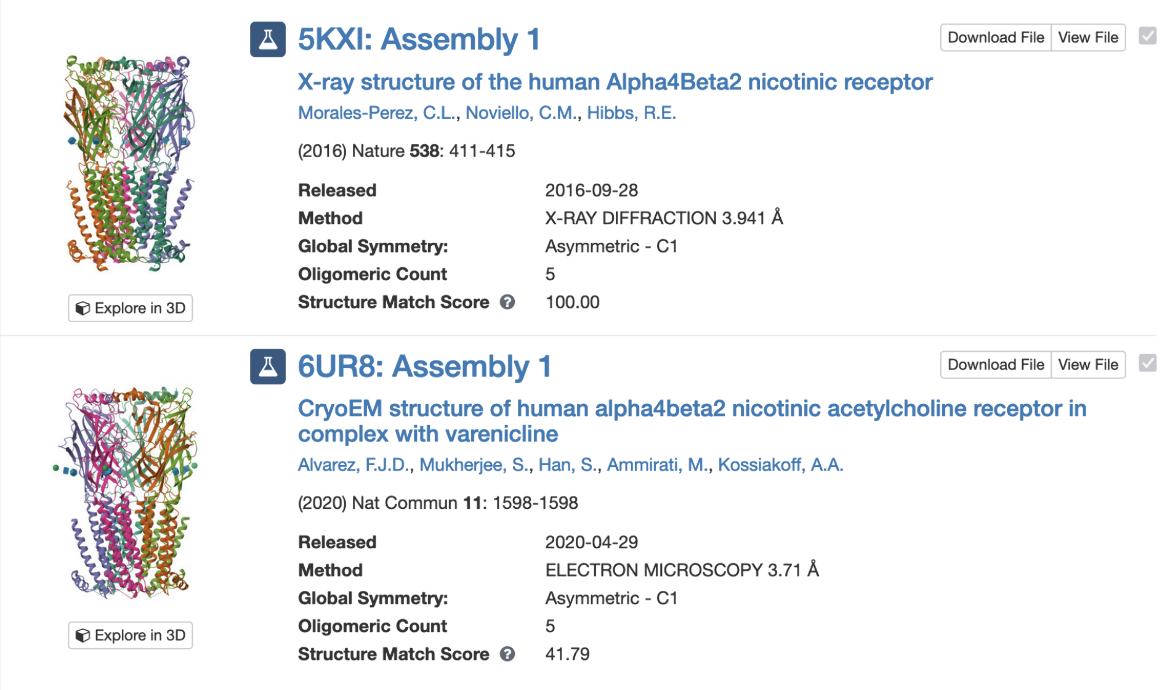
PDB entry 5kxi is the X-ray crystal structure of the human Alpha4Beta2 nicotinic receptor. The symmetry information displayed on the Structure Summary Sage is shown in Figure 6.
Note that the structure in this entry is composed of a heteropentamer. While this is classified as a global asymmetric structure, it has pseudo C5 symmetry.
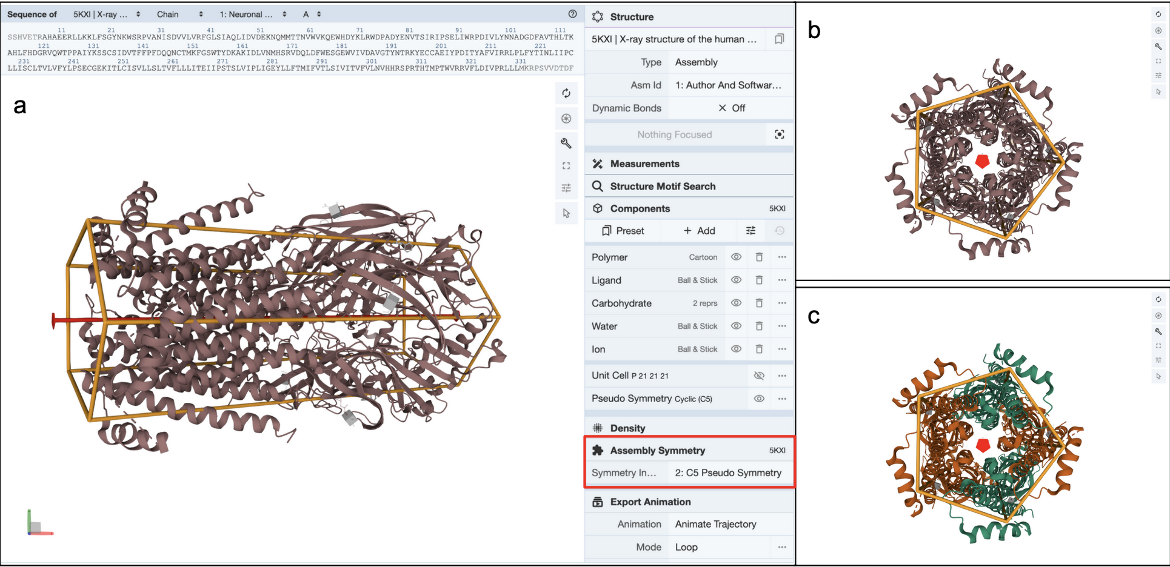
Clicking on the (3D View) link next to the “Pseudo Symmetry: Cyclic - C5” opens the structure in Mol* (Figure 7a). You can rotate the molecule to view down the 5-fold pseudosymmetry axis (Figure 7b) and color the proteins by entity type (Figure 7c). Note this assembly has a pseudo pentameric symmetry - composed of two alpha-4 (green colored) and three beta-2 (orange colored) chains.
Clicking on the “Find Similar Assemblies” on the Structure Summary Page (Figure 6) launches a strict-match assembly search to find similar assemblies in the PDB (Figure 8).
References
- Changeux, JP. (2013) 50 years of allosteric interactions: the twists and turns of the models. Nat Rev Mol Cell Biol 14, 819–829. https://doi.org/10.1038/nrm3695
- Goodsell, D. S., & Olson, A. J. (2000). Structural symmetry and protein function. Annual review of biophysics and biomolecular structure, 29, 105–153. https://doi.org/10.1146/annurev.biophys.29.1.105