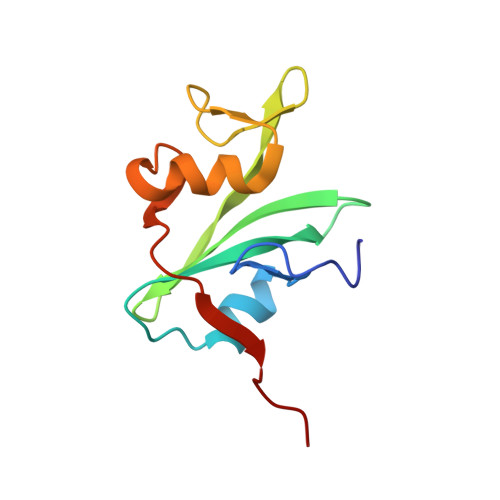
Three-dimensional solution structure of the src homology 2 domain of c-abl.
Overduin, M., Rios, C.B., Mayer, B.J., Baltimore, D., Cowburn, D.(1992) Cell 70: 697-704
- PubMed: 1505033
- DOI: https://doi.org/10.1016/0092-8674(92)90437-h
- Primary Citation of Related Structures:
1AB2 - PubMed Abstract:
SH2 regions are protein motifs capable of binding target protein sequences that contain a phosphotyrosine. The solution structure of the abl SH2 product, a protein of 109 residues and 12.1 kd, has been determined by multidimensional nuclear magnetic resonance spectroscopy. It is a compact spherical domain with a pair of three-stranded antiparallel beta sheets and a C-terminal alpha helix enclosing the hydrophobic core. Three arginines project from a short N-terminal alpha helix and one beta sheet into the putative phosphotyrosine-binding site, which lies on a face distal from the termini. Comparison with other SH2 sequences supports a common global fold and mode of phosphotyrosine binding for this family.
- Laboratories of The Rockefeller University New York, New York 10021.
Organizational Affiliation: