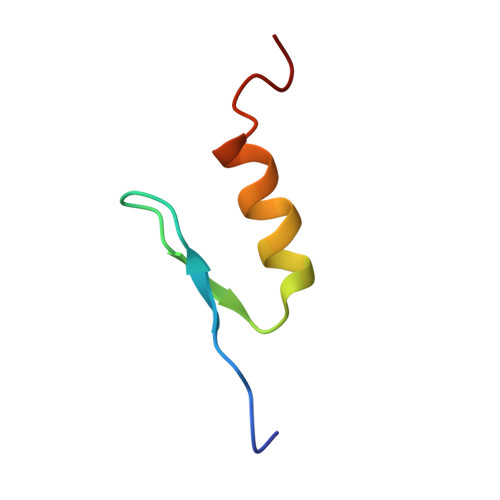
Solution structure of the transactivation domain of ATF-2 comprising a zinc finger-like subdomain and a flexible subdomain.
Nagadoi, A., Nakazawa, K., Uda, H., Okuno, K., Maekawa, T., Ishii, S., Nishimura, Y.(1999) J Mol Biology 287: 593-607
- PubMed: 10092462
- DOI: https://doi.org/10.1006/jmbi.1999.2620
- Primary Citation of Related Structures:
1BHI - PubMed Abstract:
Activating transcription factor-2 (ATF-2) is a transcription factor that binds to cAMP response element (CRE). ATF-2 contains two functional domains, an N-terminal transactivation domain and a C-terminal DNA-binding domain. The DNA-binding domain contains the basic leucine zipper (bZip) motif. Here, the three-dimensional structure of the transactivation domain of ATF-2 has been determined by NMR. The transactivation domain consists of two subdomains: the structure of an N-terminal half (N-subdomain) is well determined, while a C-terminal half (C-subdomain) takes a highly flexible and disordered structure. The architecture of the N-subdomain is very similar to that of the well-known zinc finger motif found in DNA-binding domains, consisting of an antiparallel beta-sheet and an alpha-helix. The zinc atom is tetrahedrally coordinated to two cysteine residues and two histidine residues. Amino acids that form the hydrophobic core in all of the DNA-binding zinc fingers are well conserved in the N-subdomain of the transactivation domain, whereas some amino acids that are responsible for binding to the phosphate backbone of DNA in the DNA-binding zinc fingers are substituted with other amino acids. The flexible C-subdomain, which contains two threonine residues that the stress-activated protein kinases phosphorylate, is likely to undergo a conformational change by specific binding to a target protein.
- Graduate School of Integrated Science, Yokohama City University, 22-2 Seto Kanazawa-ku, Yokohama, 236-0027, Japan.
Organizational Affiliation: