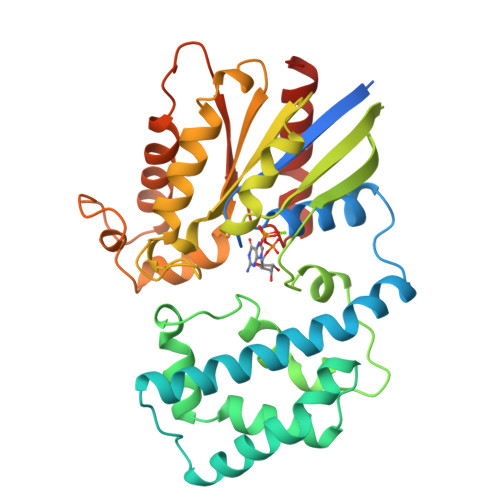
Structure of Gialpha1.GppNHp, autoinhibition in a galpha protein-substrate complex.
Coleman, D.E., Sprang, S.R.(1999) J Biological Chem 274: 16669-16672
- PubMed: 10358003
- DOI: https://doi.org/10.1074/jbc.274.24.16669
- Primary Citation of Related Structures:
1CIP - PubMed Abstract:
The structure of the G protein Gialpha1 complexed with the nonhydrolyzable GTP analog guanosine-5'-(betagamma-imino)triphosphate (GppNHp) has been determined at a resolution of 1.5 A. In the active site of Gialpha1. GppNHp, a water molecule is hydrogen bonded to the side chain of Glu43 and to an oxygen atom of the gamma-phosphate group. The side chain of the essential catalytic residue Gln204 assumes a conformation which is distinctly different from that observed in complexes with either guanosine 5'-O-3-thiotriphosphate or the transition state analog GDP.AlF4-. Hydrogen bonding and steric interactions position Gln204 such that it interacts with a presumptive nucleophilic water molecule, but cannot interact with the pentacoordinate transition state. Gln204 must be released from this auto-inhibited state to participate in catalysis. RGS proteins may accelerate the rate of GTP hydrolysis by G protein alpha subunits, in part, by inserting an amino acid side chain into the site occupied by Gln204, thereby destabilizing the auto-inhibited state of Galpha.
Organizational Affiliation:
Department of Biochemistry, The University of Texas Southwestern Medical Center, Dallas, Texas 75235-9050, USA.