Ligand Induced Conformational Transitions of Tissue Factor. Crystal Structure of the Tissue Factor:Factor VIIa Complex.
Huang, M., Ruf, W., Edgington, T.S., Wilson, I.A.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Tissue factor | A [auth T] | 210 | Homo sapiens | Mutation(s): 0 | 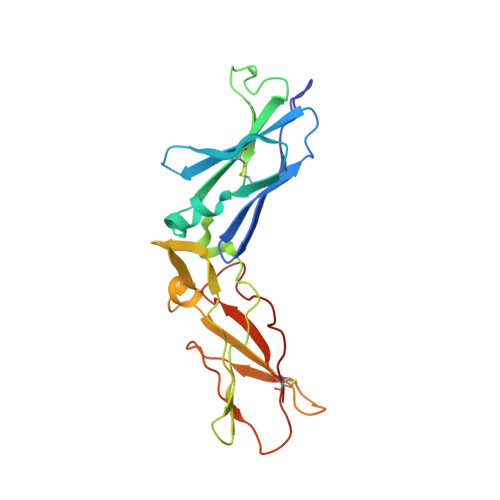 |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P13726 (Homo sapiens) Explore P13726 Go to UniProtKB: P13726 | |||||
PHAROS: P13726 GTEx: ENSG00000117525 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P13726 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| factor VIIa light chain | B [auth L] | 95 | Homo sapiens | Mutation(s): 0 EC: 3.4.21.21 | 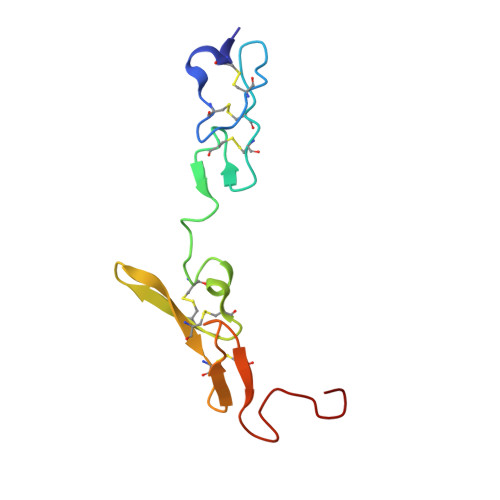 |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P08709 (Homo sapiens) Explore P08709 Go to UniProtKB: P08709 | |||||
PHAROS: P08709 GTEx: ENSG00000057593 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P08709 | ||||
Glycosylation | |||||
| Glycosylation Sites: 1 | Go to GlyGen: P08709-1 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| factor VIIa heavy chain | C [auth H] | 254 | Homo sapiens | Mutation(s): 0 EC: 3.4.21.21 | 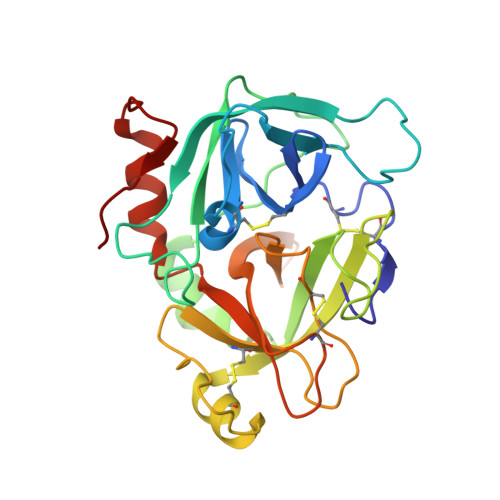 |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P08709 (Homo sapiens) Explore P08709 Go to UniProtKB: P08709 | |||||
PHAROS: P08709 GTEx: ENSG00000057593 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P08709 | ||||
Glycosylation | |||||
| Glycosylation Sites: 1 | Go to GlyGen: P08709-1 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 5 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| 0Z6 Query on 0Z6 | F [auth H] | D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-phenylalaninamide C25 H36 Cl N6 O3 ZKHBINZTIMXMQW-CLAROIROSA-O |  | ||
| NAG Query on NAG | G [auth H] | 2-acetamido-2-deoxy-beta-D-glucopyranose C8 H15 N O6 OVRNDRQMDRJTHS-FMDGEEDCSA-N |  | ||
| GLC Query on GLC | D [auth L] | alpha-D-glucopyranose C6 H12 O6 WQZGKKKJIJFFOK-DVKNGEFBSA-N |  | ||
| FUL Query on FUL | E [auth L] | beta-L-fucopyranose C6 H12 O5 SHZGCJCMOBCMKK-KGJVWPDLSA-N |  | ||
| CA Query on CA | H | CALCIUM ION Ca BHPQYMZQTOCNFJ-UHFFFAOYSA-N |  | ||
Entity ID: 6 | |||||
|---|---|---|---|---|---|
| ID | Chains | Name | Type/Class | 2D Diagram | 3D Interactions |
| PRD_000369 (0Z6) Query on PRD_000369 | F [auth H] | D-Phe-Phe-Arg Chloromethylketone | Peptide-like / Inhibitor |  | |
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 77.038 | α = 90 |
| b = 68.302 | β = 92.12 |
| c = 78.438 | γ = 90 |
| Software Name | Purpose |
|---|---|
| DENZO | data reduction |
| SCALEPACK | data scaling |
| AMoRE | phasing |
| CNS | refinement |