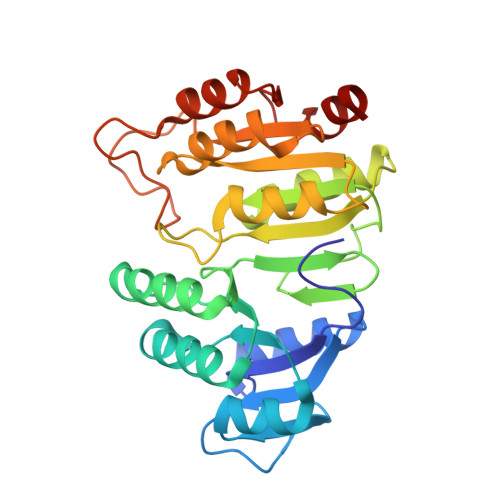
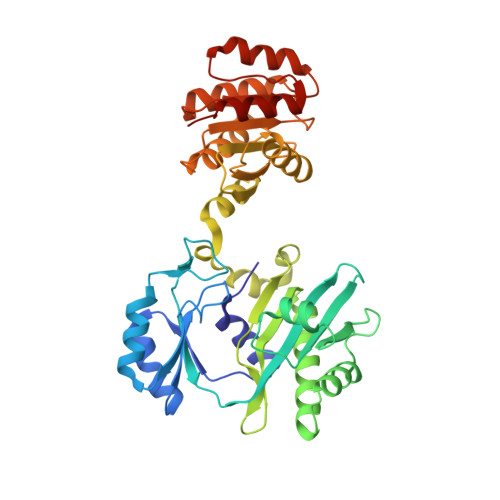
Two glutamate residues, Glu 208 alpha and Glu 197 beta, are crucial for phosphorylation and dephosphorylation of the active-site histidine residue in succinyl-CoA synthetase.
Fraser, M.E., Joyce, M.A., Ryan, D.G., Wolodko, W.T.(2002) Biochemistry 41: 537-546
- PubMed: 11781092
- DOI: https://doi.org/10.1021/bi011518y
- Primary Citation of Related Structures:
1JKJ, 1JLL - PubMed Abstract:
Succinyl-CoA synthetase catalyzes the reversible reaction succinyl-CoA + NDP + P(i) <--> succinate + CoA + NTP (N denoting adenosine or guanosine). The enzyme consists of two different subunits, designated alpha and beta. During the reaction, a histidine residue of the alpha-subunit is transiently phosphorylated. This histidine residue interacts with Glu 208 alpha at site I in the structures of phosphorylated and dephosphorylated Escherichia coli SCS. We postulated that Glu 197 beta, a residue in the nucleotide-binding domain, would provide similar stabilization of the histidine residue during the actual phosphorylation/dephosphorylation by nucleotide at site II. In this work, these two glutamate residues have been mutated individually to aspartate or glutamine. Glu 197 beta has been additionally mutated to alanine. The mutant proteins were tested for their ability to be phosphorylated in the forward or reverse direction. The aspartate mutant proteins can be phosphorylated in either direction, while the E208 alpha Q mutant protein can only be phosphorylated by NTP, and the E197 beta Q mutant protein can only be phosphorylated by succinyl-CoA and P(i). These results demonstrate that the length of the side chain at these positions is not critical, but that the charge is. Most significantly, the E197 beta A mutant protein could not be phosphorylated in either direction. Its crystal structure shows large differences from the wild-type enzyme in the conformation of two residues of the alpha-subunit, Cys 123 alpha-Pro 124 alpha. We postulate that in this conformation, the protein cannot productively bind succinyl-CoA for phosphorylation via succinyl-CoA and P(i).
- Department of Biochemistry, University of Western Ontario, London, Ontario, Canada N6A 5C1. mfraser4@uwo.ca
Organizational Affiliation: