NMR Solution Structure of ATTp, an Arabidopsis thaliana Trypsin Inhibitor
Zhao, Q., Chae, Y.K., Markley, J.L.(2002) Biochemistry 41: 12284-12296
- PubMed: 12369816
- DOI: https://doi.org/10.1021/bi025702a
- Primary Citation of Related Structures:
1JXC - PubMed Abstract:
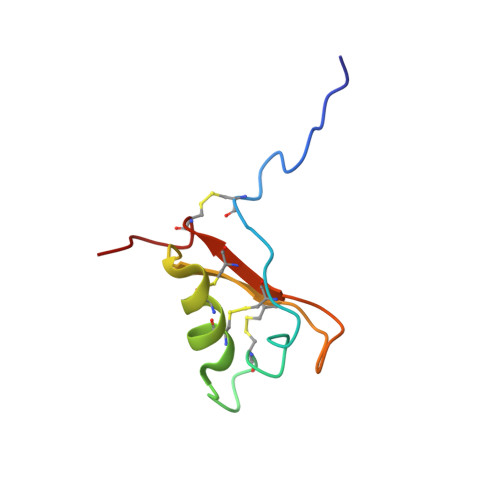
The three-dimensional structure of the precursor form of the Arabidopsis thaliana trypsin inhibitor (ATT(p), GenBank entry Z46816), a 68-residue (approximately 7.5 kDa) rapeseed class proteinase inhibitor, has been determined in solution at pH 5.0 and 25 degrees C by multinuclear magnetic resonance spectroscopy. The protein contains one alpha-helix and two strands of antiparallel beta-sheet, with a type IV beta-turn connecting the two strands. The alpha-helix and the inhibitory loop are connected to the beta-sheet through three disulfide bridges; a fourth disulfide bridge connects the N- and C-termini. The overall structural topology of ATT(p) is similar to those of the sweet tasting protein brazzein (rmsd of 3.0 A) and the antifungal protein Rs-Afp1 [a knottin protein from radish (Raphanus sativus), rmsd of 2.7 A]. The precursor segment in ATT(p) is disordered, as visualized by the final 20-conformer ensemble and as confirmed by (15)N heteronuclear NOE analysis. The overall fold of ATT(p) is distinct from those of other classes of serine proteinase inhibitors except in the inhibitor loop; therefore, it represents a new inhibitor fold.
- Department of Biochemistry, University of Wisconsin-Madison, 53706, USA.
Organizational Affiliation: