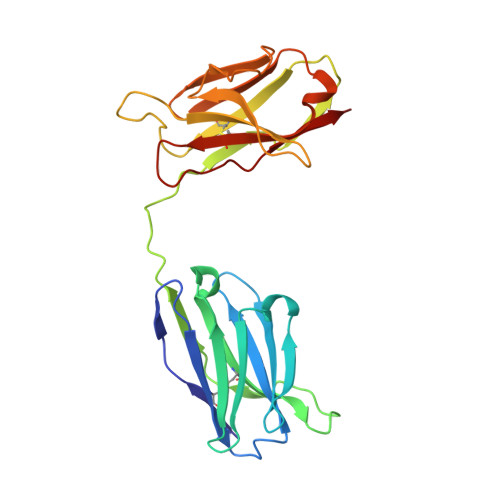
Crystal structure of a diabody, a bivalent antibody fragment.
Perisic, O., Webb, P.A., Holliger, P., Winter, G., Williams, R.L.(1994) Structure 2: 1217-1226
- PubMed: 7704531
- DOI: https://doi.org/10.1016/s0969-2126(94)00123-5
- Primary Citation of Related Structures:
1LMK - PubMed Abstract:
Diabodies are dimeric antibody fragments. In each polypeptide, a heavy-chain variable domain (VH) is linked to a light-chain variable domain (VL) but unlike single-chain Fv fragments, each antigen-binding site is formed by pairing of one VH and one VL domain from the two different polypeptides. Diabodies thus have two antigen-binding sites, and can be bispecific. Direct structural evidence is lacking for the connections and dimeric interactions between the two polypeptides of the diabody. The 2.6 A resolution structure has been determined for a bivalent diabody with a flexible five-residue polypeptide linker between the (amino-terminal) VH and (carboxy-terminal) VL domains. The asymmetric unit of the crystal consists of four polypeptides comprising two diabodies; for one of these polypeptides the linker can be traced between the VH and VL domains. Within each diabody the two associated VH and VL domains make back-to-back interactions through the VH domains, and there is an extensive VL-VL interface between the two diabodies in the asymmetric unit. The structure of the diabody is very similar to that which had been predicted by molecular modelling. Diabodies directed against cell-surface antigens should be capable of bringing together two cells, such as in cell-targeted therapy, because the two antigen-binding sites of the diabody are at opposite ends of the molecule and separated by approximately 65 A.
- Centre for Protein Engineering, MRC Centre, Cambridge, UK.
Organizational Affiliation: