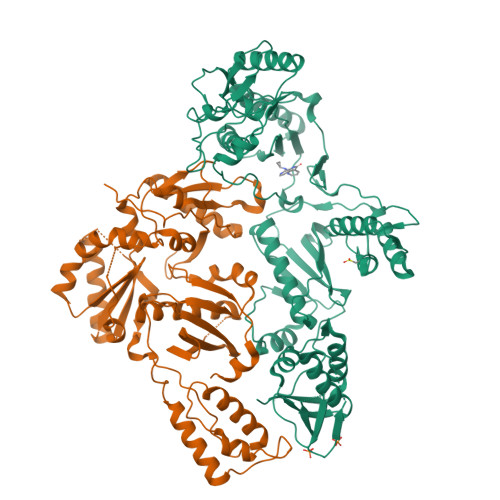
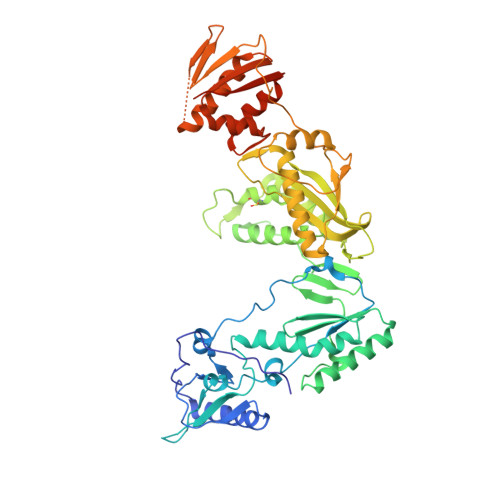
Crystal structures of Zidovudine- or Lamivudine-resistant human immunodeficiency virus type 1 reverse transcriptases containing mutations at codons 41, 184, and 215.
Chamberlain, P.P., Ren, J., Nichols, C.E., Douglas, L., Lennerstrand, J., Larder, B.A., Stuart, D.I., Stammers, D.K.(2002) J Virol 76: 10015-10019
- PubMed: 12208978
- DOI: https://doi.org/10.1128/jvi.76.19.10015-10019.2002
- Primary Citation of Related Structures:
1LW0, 1LW2, 1LWC, 1LWE, 1LWF - PubMed Abstract:
Six structures of human immunodeficiency virus type 1 (HIV-1) reverse transcriptase (RT) containing combinations of resistance mutations for zidovudine (AZT) (M41L and T215Y) or lamivudine (M184V) have been determined as inhibitor complexes. Minimal conformational changes in the polymerase or nonnucleoside RT inhibitor sites compared to the mutant RTMC (D67N, K70R, T215F, and K219N) are observed, indicating that such changes may occur only with certain combinations of mutations. Model building M41L and T215Y into HIV-1 RT-DNA and docking in ATP that is utilized in the pyrophosphorolysis reaction for AZT resistance indicates that some conformational rearrangement appears necessary in RT for ATP to interact simultaneously with the M41L and T215Y mutations.
Organizational Affiliation:
Division of Structural Biology, The Wellcome Trust Centre for Human Genetics, University of Oxford, Oxford OX3 7BN, United Kingdom.