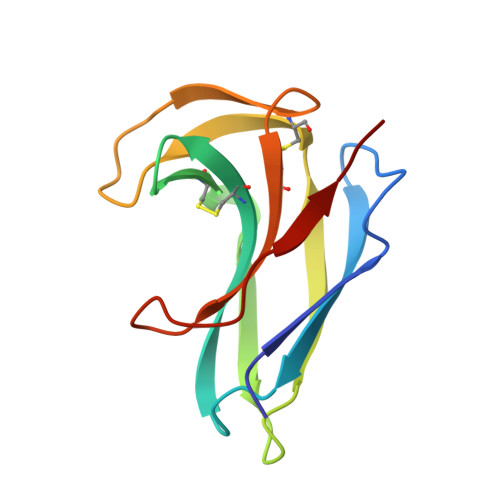Crystal structure of apo-neocarzinostatin at 0.15-nm resolution.
Teplyakov, A., Obmolova, G., Wilson, K., Kuromizu, K.(1993) Eur J Biochem 213: 737-741
- PubMed: 8477746
- DOI: https://doi.org/10.1111/j.1432-1033.1993.tb17814.x
- Primary Citation of Related Structures:
1NOA - PubMed Abstract:
The three-dimensional structure of apo-neocarzinostatin, an antitumour antibiotic protein isolated from Streptomyces carzinostaticus, has been determined by X-ray diffraction at 0.15-nm resolution and refined to R = 17.2%. The crystal structure of neocarzinostatin is similar to that of the related proteins actinoxanthin and macromomycin. It is also in good agreement with the solution structure determined by NMR spectroscopy. The protein molecule consists of a seven-stranded antiparallel beta-sandwich and a smaller lobe formed by two beta-ribbons. A deep cleft between the two lobes is a putative chromophore binding site. Side chains of Trp39, Leu45, Phe52, Phe78 and the disulphide Cys37-Cys47 aligning the binding cleft in neocarzinostatin suggest the importance of hydrophobic interactions in stabilizing the chromophore molecule. Comparison of the atomic models of neocarzinostatin, actinoxanthin and macromomycin reveals functional residues which might determine specificity towards different chromophores.
- EMBL, c/o DESY, Hamburg, Germany.
Organizational Affiliation: