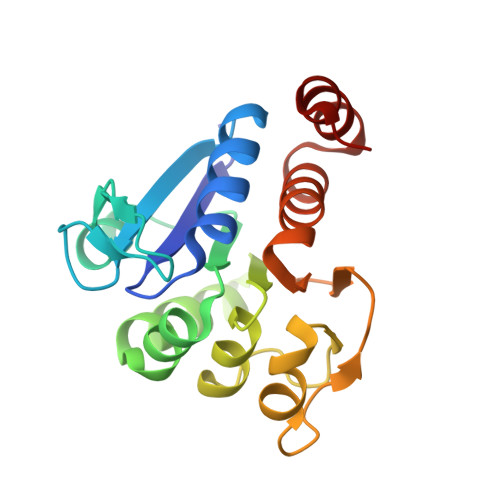
The 1.1 A resolution crystal structure of DJ-1, the protein mutated in autosomal recessive early onset Parkinson's disease
Wilson, M.A., Collins, J.L., Hod, Y., Ringe, D., Petsko, G.A.(2003) Proc Natl Acad Sci U S A 100: 9256-9261
- PubMed: 12855764
- DOI: https://doi.org/10.1073/pnas.1133288100
- Primary Citation of Related Structures:
1P5F - PubMed Abstract:
Mutations in DJ-1, a human gene with homologues in organisms from all kingdoms of life, have been shown to be associated with autosomal recessive, early onset Parkinson's disease (PARK7). We report here the three-dimensional structure of the DJ-1 protein, determined at a resolution of 1.1 A by x-ray crystallography. The chain fold of DJ-1 resembles those of a bacterial protein, PfpI, that has been annotated as a cysteine protease, and of a domain of a bacterial catalase whose role in the activity of that enzyme is uncertain. In contrast to PfpI, a hexameric protein whose oligomeric structure is essential for its putative proteolytic activity, DJ-1 is a dimer with completely different intersubunit contacts. The proposed catalytic triad of PfpI is absent from the corresponding region of the structure of DJ-1, and biochemical assays fail to detect any protease activity for purified DJ-1. A highly conserved cysteine residue, which is catalytically essential in homologues of DJ-1, shows an extreme sensitivity to radiation damage and may be subject to other forms of oxidative modification as well. The structure suggests that the loss of function caused by the Parkinson's-associated mutation L166P in DJ-1 is due to destabilization of the dimer interface. Taken together, the crystal structure of human DJ-1 plus other observations suggest the possible involvement of this protein in the cellular oxidative stress response and a general etiology of neurodegenerative diseases.
- Department of Biochemistry and Chemistry and Rosenstiel Basic Medical Sciences Research Center, Brandeis University, 415 South Street, MS 029, Waltham, MA 02454-9110, USA.
Organizational Affiliation: