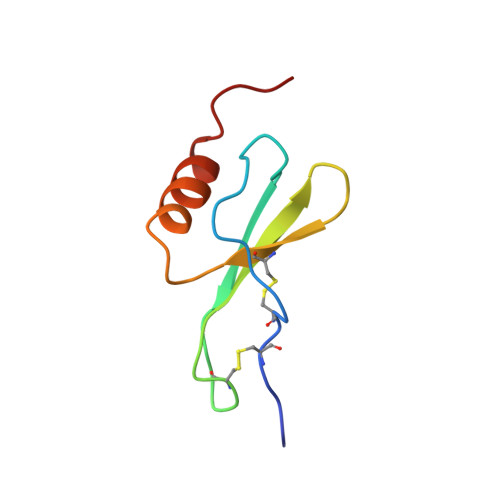
Nuclear magnetic resonance solution structure of truncated human GRObeta [5-73] and its structural comparison with CXC chemokine family members GROalpha and IL-8.
Qian, Y.Q., Johanson, K.O., McDevitt, P.(1999) J Mol Biology 294: 1065-1072
- PubMed: 10600366
- DOI: https://doi.org/10.1006/jmbi.1999.3333
- Primary Citation of Related Structures:
1QNK - PubMed Abstract:
The three-dimensional structure of a novel four amino acid truncated form of the CXC chemokine GRObeta [5-73] isolated from bone marrow stromal cells with potent hematopoietic and anti-infective activities has been determined by two-dimensional (1)H nuclear magnetic resonance (NMR) spectroscopy in solution. On the basis of 1878 upper distance constraints derived from nuclear Overhauser effects (NOE) and 314 dihedral angle constraints, a group of 20 conformers representing the solution structure of the human GRObeta [5-73] was computed with the program DYANA. At the concentrations used for NMR study, GRObeta [5-73] forms a dimer in solution that is architectured by a six-stranded antiparallel beta-sheet (residues 25 to 29, 39 to 44, 49 to 52) and a pair of helices (residues 58 to 68) with 2-fold symmetry, while the C terminus of the protein is disordered. The average of the pairwise root-mean-square deviations of individual NMR conformers relative to the mean coordinates for the backbone atoms N, C(alpha) and C' of residues 5 to 68 is 0.47 A. Overall, the global fold of GRObeta [5-73] is similar to that of the previously reported NMR structure of GROalpha and the NMR and X-ray structures of interleukin-8. Among these three CXC chemokines, GRObeta [5-73] is most similar in structure to GROalpha. Significant differences between GRObeta [5-73], GROalpha and interleukin-8 are in the N-terminal loop comprising residues 12 to 19. The N-terminal arm containing the conserved ELR motif and the loop of residues 30 to 38 containing the GPH motif are different among these three CXC chemokines. The structural differences in these two regions may be responsible for the specificity of the receptor binding and biological activity of different chemokines.
- Department of Physical & Structural Chemistry, SmithKline Beecham Pharmaceuticals, King of Prussia, PA 19406, USA. Yanqiu_Qian@sbphrd.com
Organizational Affiliation: