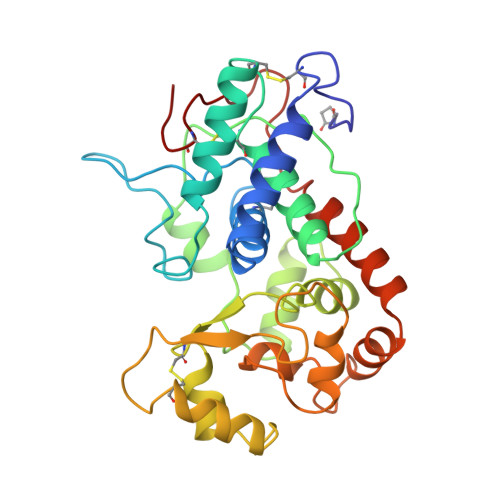
The crystal structure of peanut peroxidase.
Schuller, D.J., Ban, N., Huystee, R.B., McPherson, A., Poulos, T.L.(1996) Structure 4: 311-321
- PubMed: 8805539
- DOI: https://doi.org/10.1016/s0969-2126(96)00035-4
- Primary Citation of Related Structures:
1SCH - PubMed Abstract:
Peroxidases catalyze a wide variety of peroxide-dependent oxidations. Based on sequence alignments, heme peroxidases have been divided into three classes. Crystal structures are available for peroxidases of classes I and II, but until now no structure has been determined for class III, the classical extracellular plant peroxidases. The crystal structure of peanut peroxidase has been solved to 2.7 A resolution. The helical fold is similar to that of known peroxidase structures. The 294-residue polypeptide chain is accompanied by a heme and two calcium ions, and there is some evidence of glycosylation. This is the first complete structure of a class III peroxidase and as such should serve as a model for other class III enzymes including the much-studied horseradish peroxidase. It may also aid in the interpretation of functional differences between the peroxidase classes. Ten helices conserved in class I and II peroxidases are also found in peanut peroxidase. Key residues of the heme environment and the location of two calcium ions are shared with class II peroxidases. Peanut peroxidase contains three unique helices, two of which contribute to the substrate access channel leading to the heme edge.
- Department of Molecular Biology & Biochemistry, University of California, Irvine, CA 92717-3900, USA.
Organizational Affiliation: