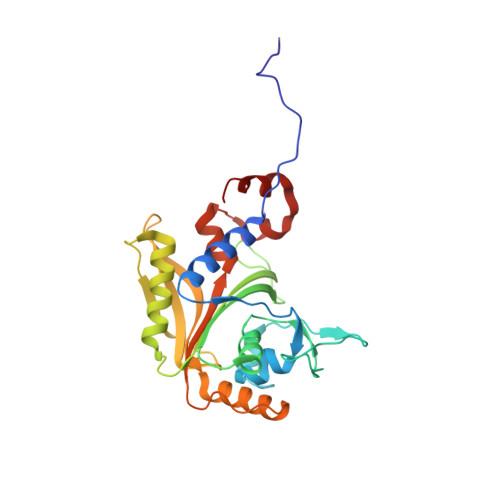
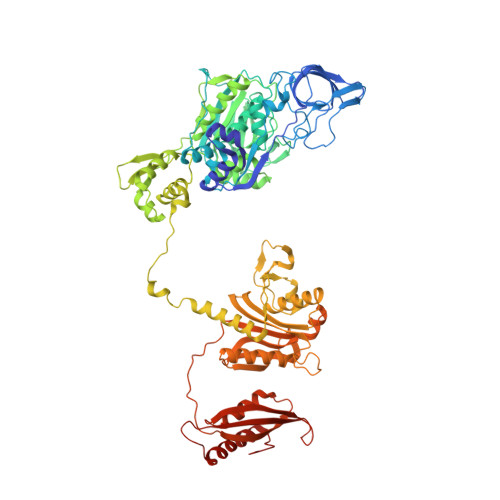
Structural Basis for Discrimination of L-Phenylalanine from L-Tyrosine by Phenylalanyl-tRNA Synthetase
Kotik-Kogan, O.M., Moor, N.A., Tworowski, D.E., Safro, M.G.(2005) Structure 13: 1799-1807
- PubMed: 16338408
- DOI: https://doi.org/10.1016/j.str.2005.08.013
- Primary Citation of Related Structures:
2AKW, 2ALY, 2AMC - PubMed Abstract:
Aminoacyl-tRNA synthetases (aaRSs) exert control over the faithful transfer of amino acids onto cognate tRNAs. Since chemical structures of various amino acids closely resemble each other, it is difficult to discriminate between them. Editing activity has been evolved by certain aaRSs to resolve the problem. In this study, we determined the crystal structures of complexes of T. thermophilus phenylalanyl-tRNA synthetase (PheRS) with L-tyrosine, p-chloro-phenylalanine, and a nonhydrolyzable tyrosyl-adenylate analog. The structures demonstrate plasticity of the synthetic site capable of binding substrates larger than phenylalanine and provide a structural basis for the proofreading mechanism. The editing site is localized at the B3/B4 interface, 35 A from the synthetic site. Glubeta334 plays a crucial role in the specific recognition of the Tyr moiety in the editing site. The tyrosyl-adenylate analog binds exclusively in the synthetic site. Both structural data and tyrosine-dependent ATP hydrolysis enhanced by tRNA(Phe) provide evidence for a preferential posttransfer editing pathway in the phenylalanine-specific system.
- Department of Structural Biology, Weizmann Institute of Science, 76100 Rehovot, Israel.
Organizational Affiliation: