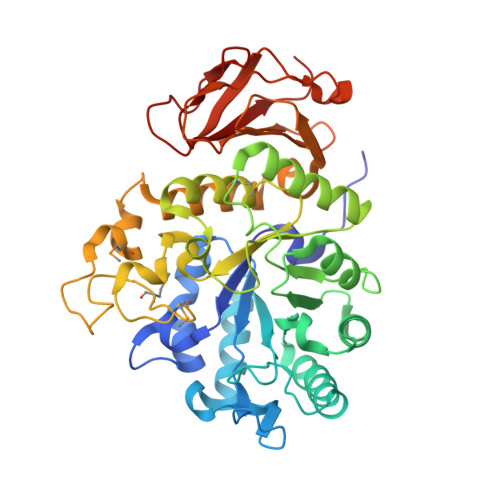
The structure of human 4F2hc ectodomain provides a model for homodimerization and electrostatic interaction with plasma membrane.
Fort, J., de la Ballina, L.R., Burghardt, H.E., Ferrer-Costa, C., Turnay, J., Ferrer-Orta, C., Uson, I., Zorzano, A., Fernandez-Recio, J., Orozco, M., Lizarbe, M.A., Fita, I., Palacin, M.(2007) J Biological Chem 282: 31444-31452
- PubMed: 17724034
- DOI: https://doi.org/10.1074/jbc.M704524200
- Primary Citation of Related Structures:
2DH2, 2DH3 - PubMed Abstract:
4F2hc (CD98hc) is a multifunctional type II membrane glycoprotein involved in amino acid transport and cell fusion, adhesion, and transformation. The structure of the ectodomain of human 4F2hc has been solved using monoclinic (Protein Data Bank code 2DH2) and orthorhombic (Protein Data Bank code 2DH3) crystal forms at 2.1 and 2.8 A, respectively. It is composed of a (betaalpha)(8) barrel and an antiparallel beta(8) sandwich related to bacterial alpha-glycosidases, although lacking key catalytic residues and consequently catalytic activity. 2DH3 is a dimer with Zn(2+) coordination at the interface. Human 4F2hc expressed in several cell types resulted in cell surface and Cys(109) disulfide bridge-linked homodimers with major architectural features of the crystal dimer, as demonstrated by cross-linking experiments. 4F2hc has no significant hydrophobic patches at the surface. Monomer and homodimer have a polarized charged surface. The N terminus of the solved structure, including the position of Cys(109) residue located four residues apart from the transmembrane domain, is adjacent to the positive face of the ectodomain. This location of the N terminus and the Cys(109)-intervening disulfide bridge imposes space restrictions sufficient to support a model for electrostatic interaction of the 4F2hc ectodomain with membrane phospholipids. These results provide the first crystal structure of heteromeric amino acid transporters and suggest a dynamic interaction of the 4F2hc ectodomain with the plasma membrane.
- Barcelona Science Park and the Department of Biochemistry and Molecular Biology, Faculty of Biology, University of Barcelona and Centro de Investigación Biomédica en Red de Enfermedades Raras, E-08028 Barcelona, Spain.
Organizational Affiliation: