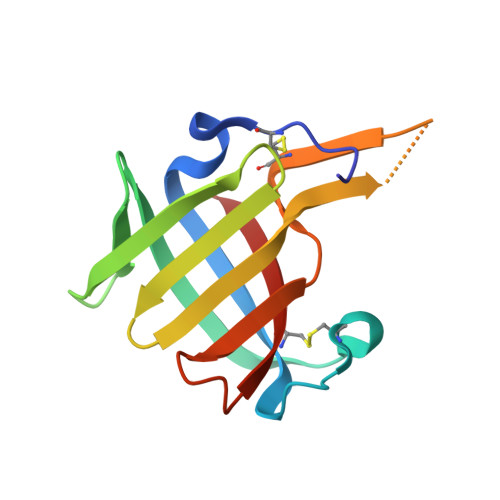
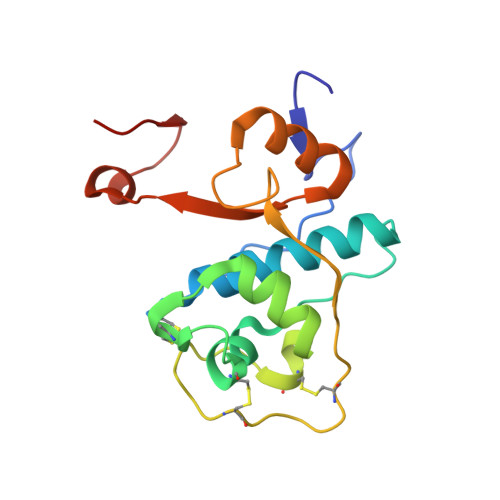
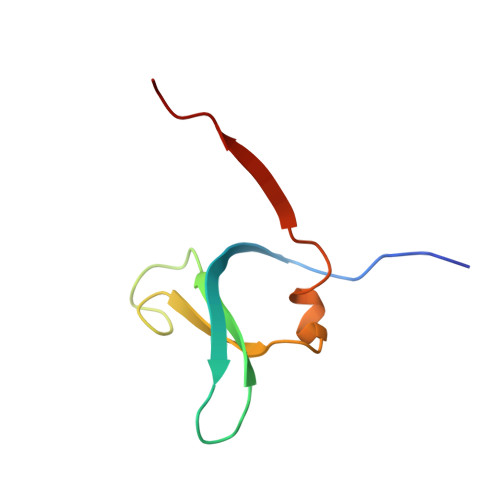
The crystal structure of human dipeptidyl peptidase I (cathepsin C) in complex with the inhibitor Gly-Phe-CHN2
Molgaard, A., Arnau, J., Lauritzen, C., Larsen, S., Petersen, G., Pedersen, J.(2007) Biochem J 401: 645-650
- PubMed: 17020538
- DOI: https://doi.org/10.1042/BJ20061389
- Primary Citation of Related Structures:
2DJF, 2DJG - PubMed Abstract:
hDDPI (human dipeptidyl peptidase I) is a lysosomal cysteine protease involved in zymogen activation of granule-associated proteases, including granzymes A and B from cytotoxic T-lymphocytes and natural killer cells, cathepsin G and neutrophil elastase, and mast cell tryptase and chymase. In the present paper, we provide the first crystal structure of an hDPPI-inhibitor complex. The inhibitor Gly-Phe-CHN2 (Gly-Phe-diazomethane) was co-crystallized with hDPPI and the structure was determined at 2.0 A (1 A=0.1 nm) resolution. The structure of the native enzyme was also determined to 2.05 A resolution to resolve apparent discrepancies between the complex structure and the previously published structure of the native enzyme. The new structure of the native enzyme is, within the experimental error, identical with the structure of the enzyme-inhibitor complex presented here. The inhibitor interacts with three subunits of hDPPI, and is covalently bound to Cys234 at the active site. The interaction between the totally conserved Asp1 of hDPPI and the ammonium group of the inhibitor forms an essential interaction that mimics enzyme-substrate interactions. The structure of the inhibitor complex provides an explanation of the substrate specificity of hDPPI, and gives a background for the design of new inhibitors.
- Centre for Crystallographic Studies, University of Copenhagen, Universitetsparken 5, DK-2100 Copenhagen Ø, Denmark. anne@ccs.ki.ku.dk
Organizational Affiliation: