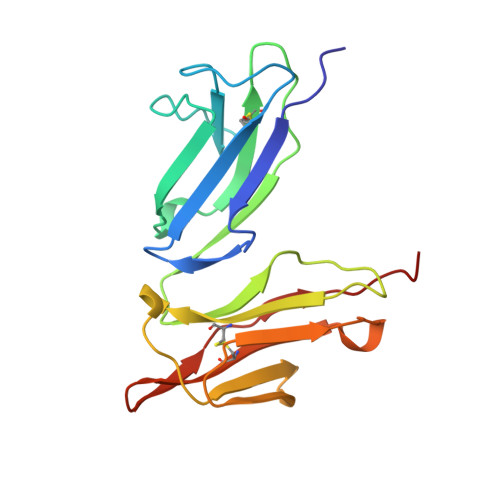
Crystal structure of the soluble form of the human fcgamma-receptor IIb: a new member of the immunoglobulin superfamily at 1.7 A resolution.
Sondermann, P., Huber, R., Jacob, U.(1999) EMBO J 18: 1095-1103
- PubMed: 10064577
- DOI: https://doi.org/10.1093/emboj/18.5.1095
- Primary Citation of Related Structures:
2FCB - PubMed Abstract:
Fcgamma-receptors (FcgammaRs) represent the link between the humoral and cellular immune responses. Via the binding to FcgammaR-positive cells, immunocomplexes trigger several functions such as endocytosis, antibody-dependent cell-mediated cytotoxity (ADCC) and the release of mediators, making them a valuable target for the modulation of the immune system. We solved the crystal structure of the soluble human Fcgamma-receptor IIb (sFcgammaRIIb) to 1.7 A resolution. The structure reveals two typical immunoglobulin (Ig)-like domains enclosing an angle of approximately 70 degrees, leading to a heart-shaped overall structure. In contrast to the observed flexible arrangement of the domains in other members of the Ig superfamily, the two domains are anchored by several hydrogen bonds. The structure reveals that the residues relevant for IgG binding, which were already partially characterized by mutagenesis studies, are located within the BC, C'E and FG loops between the beta-strands of the second domain. Moreover, we discuss a model for the sFcgammaRIIb:IgG complex. In this model, two FcgammaR molecules bind one IgG molecule with their second domains, while the first domain points away from the complex and is therefore available for binding other cell surface molecules, by which potential immunosuppressing functions could be mediated.
- Max-Planck-Institut für Biochemie, Abteilung Strukturforschung, Am Klopferspitz 18a, D-82152 Martinsried, Germany.
Organizational Affiliation: