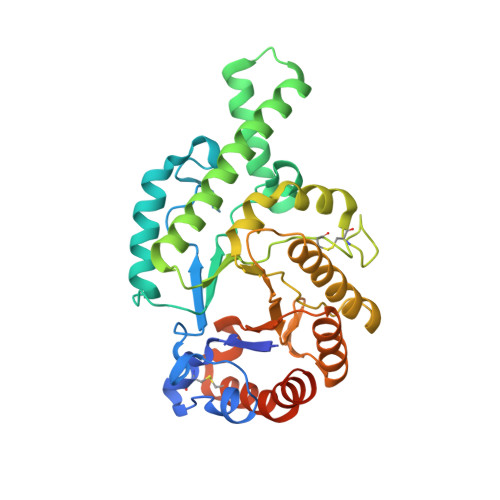
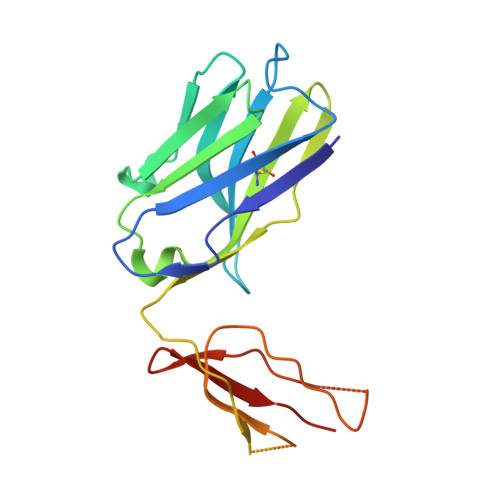
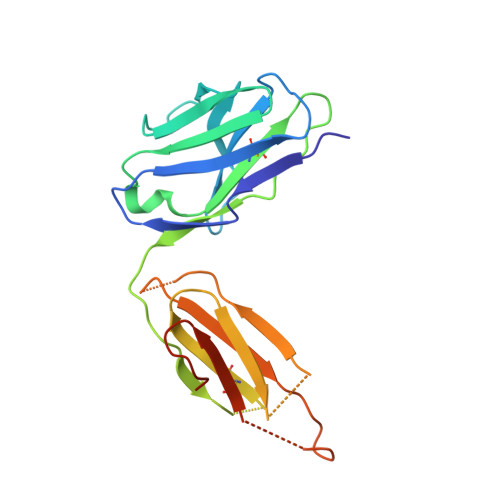
Identification of a B-Cell Epitope of Hyaluronidase, a Major Bee Venom Allergen, from its Crystal Structure in Complex with a Specific Fab.
Padavattan, S., Schirmer, T., Schmidt, M., Akdis, C., Valenta, R., Mittermann, I., Soldatova, L., Slater, J., Mueller, U., Markovic-Housley, Z.(2007) J Mol Biology 368: 742
- PubMed: 17374540
- DOI: https://doi.org/10.1016/j.jmb.2007.02.036
- Primary Citation of Related Structures:
2J88 - PubMed Abstract:
The major allergens of honeybee venom, hyaluronidase (Hyal) and phospholipase A2, can induce life-threatening IgE-mediated allergic reactions in humans. Although conventional immunotherapy is effective, up to 40% of patients develop allergic side effects including anaphylaxis and thus, there is a need for an improved immunotherapy. A murine monoclonal anti-Hyal IgG1 antibody (mAb 21E11), that competed for Hyal binding with IgEs from sera of bee venom allergic patients, was raised. The fragment of these IgG antibodies which bind to antigen (Fab) was produced and complexed (1:1) with Hyal. The crystal structure determination of Hyal/Fab 21E11 complex (2.6 A) enabled the identification of the Hyal-IgG interface which provides indirect information on the Hyal-IgE interaction (B-cell epitope). The epitope is composed of a linear array of nine residues (Arg138, His141-Arg148) located at the tip of a helix-turn-helix motive which protrudes away from the globular core and fits tightly into the deep surface pocket formed by the residues from the six complementarity determining regions (CDRs) of the Fab. The epitope is continuous and yet its conformation appears to be essential for Ab recognition, since the synthetic 15-mer peptide comprising the entire epitope (Arg138-Glu152) is neither recognized by mAb 21E11 nor by human IgEs. The structure of the complex provides the basis for the rational design of Hyal derivatives with reduced allergenic activity, which could be used in the development of safer allergen-specific immunotherapy.
- Division of Structural Biology, Biozentrum, University of Basel, CH-4056 Basel, Switzerland.
Organizational Affiliation: