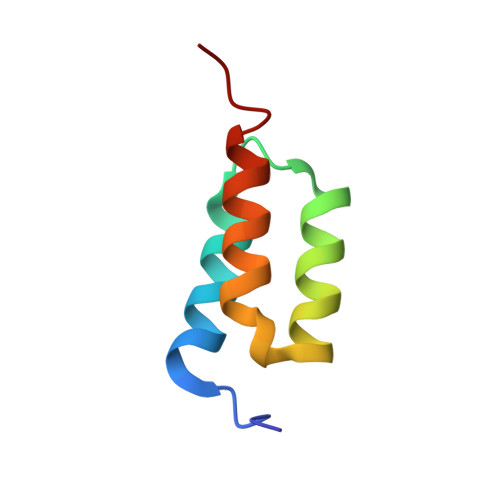
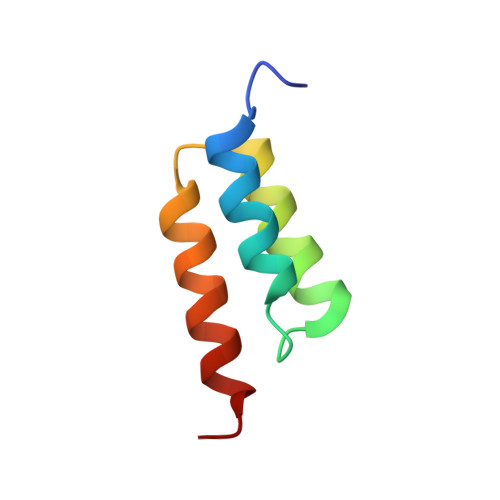
High-affinity binding to staphylococcal protein A by an engineered dimeric Affibody molecule.
Lindborg, M., Dubnovitsky, A., Olesen, K., Bjorkman, T., Abrahmsen, L., Feldwisch, J., Hard, T.(2013) Protein Eng Des Sel 26: 635-644
- PubMed: 23924760
- DOI: https://doi.org/10.1093/protein/gzt038
- Primary Citation of Related Structures:
2M5A - PubMed Abstract:
Affibody molecules are engineered binding proteins, in which the three-helix bundle motif of the Z domain derived from protein A is used as a scaffold for sequence variation. We used phage display to select Affibody binders to staphylococcal protein A itself. The best binder, called ZpA963, binds with similar affinity and kinetics to the five homologous E, D, A, B and C domains of protein A, and to a five-domain protein A construct with an average dissociation constant, K(D), of ~20 nM. The structure of ZpA963 in complex with the Z domain shows that it interacts with a surface on Z that is identical in the five protein A domains, which explains the multi-domain affinity. This property allows for high-affinity binding by dimeric Affibody molecules that simultaneously engage two protein A domains in a complex. We studied two ZpA963 dimers in which the subunits were linked by a C-terminal disulfide in a symmetric dimer or head-to-tail in a fusion protein, respectively. The dimers both bind protein A with high affinity, very slow off-rates and with saturation-dependent kinetics that can be understood in terms of dimer binding to multiple sites. The head-to-tail (ZpA963)2htt dimer binds with an off-rate of k(off) ≤ 5 × 10(-6) s(-1) and an estimated K(D) ≤ 16 pM. The results illustrate how dimers of selected monomer binding proteins can provide an efficient route for engineering of high-affinity binders to targets that contain multiple homologous domains or repeated structural units.
- Affibody AB, Gunnar Asplunds Allé 24, SE-171 63 Solna, Sweden.
Organizational Affiliation: