Structural venomics reveals evolution of a complex venom by duplication and diversification of an ancient peptide-encoding gene.
Pineda, S.S., Chin, Y.K., Undheim, E.A.B., Senff, S., Mobli, M., Dauly, C., Escoubas, P., Nicholson, G.M., Kaas, Q., Guo, S., Herzig, V., Mattick, J.S., King, G.F.(2020) Proc Natl Acad Sci U S A 117: 11399-11408
- PubMed: 32398368
- DOI: https://doi.org/10.1073/pnas.1914536117
- Primary Citation of Related Structures:
2N6N, 2N6R, 2N8K, 6BA3 - PubMed Abstract:
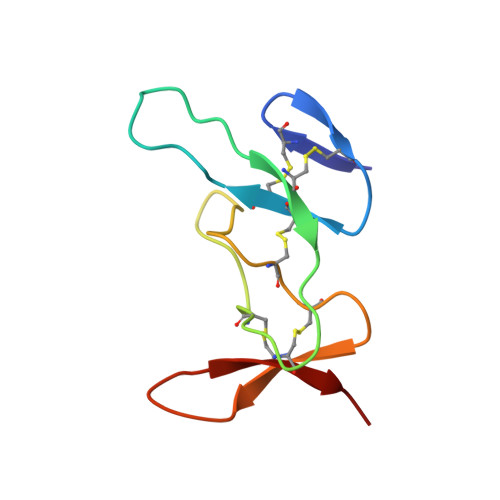
Spiders are one of the most successful venomous animals, with more than 48,000 described species. Most spider venoms are dominated by cysteine-rich peptides with a diverse range of pharmacological activities. Some spider venoms contain thousands of unique peptides, but little is known about the mechanisms used to generate such complex chemical arsenals. We used an integrated transcriptomic, proteomic, and structural biology approach to demonstrate that the lethal Australian funnel-web spider produces 33 superfamilies of venom peptides and proteins. Twenty-six of the 33 superfamilies are disulfide-rich peptides, and we show that 15 of these are knottins that contribute >90% of the venom proteome. NMR analyses revealed that most of these disulfide-rich peptides are structurally related and range in complexity from simple to highly elaborated knottin domains, as well as double-knot toxins, that likely evolved from a single ancestral toxin gene.
- Institute for Molecular Bioscience, The University of Queensland, St Lucia, QLD 4072, Australia; sandy.spineda@gmail.com glenn.king@imb.uq.edu.au.
Organizational Affiliation: