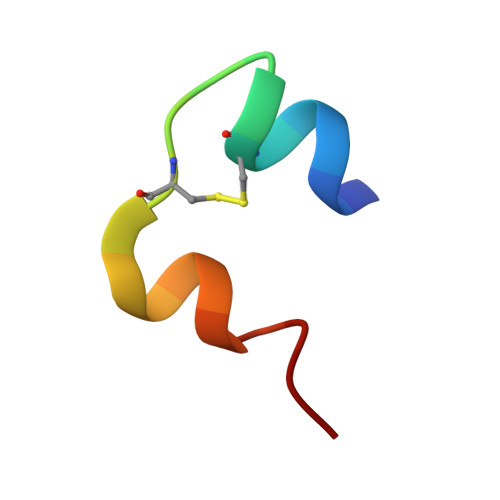
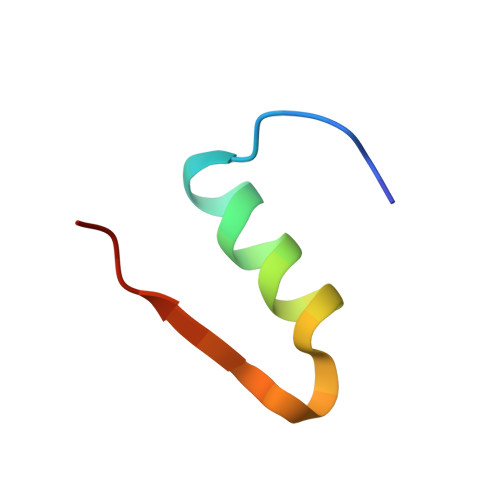
Crystal Structure of Ultralente-A Microcrystalline Insulin Suspension.
Wagner, A., Diez, J., Schulze-Briese, C., Schluckebier, G.(2009) Proteins 74: 1018
- PubMed: 18767151
- DOI: https://doi.org/10.1002/prot.22213
- Primary Citation of Related Structures:
2VJZ, 2VK0 - PubMed Abstract:
Ultralente insulin has been one of the commercially most important insulin preparations in diabetes treatment over the last 50 years. It is a suspension of insulin microcrystals which dissolve slowly following subcutaneous injection. Because of the small crystal size of about 25 x 25 x 5 microm(3) the atomic structure has been elusive until now. Here we present the crystal structures from Ultralente and their precursor microcrystals from the industrial manufacturing process. During this process insulin undergoes a conformational change within the microcrystals. Both structures show canonical folding of the insulin molecules but exhibit a number of new features when compared with other insulin structures. Surprisingly, we found that the Ultralente crystals bind the conservation agent methylparaben, which slows down dissolution of the crystals and thus contributes to the long duration of action.
- Swiss Light Source, Paul Scherrer Institute, CH-5323 Villigen PSI, Switzerland. armin.wagner@diamond.ac.uk
Organizational Affiliation: