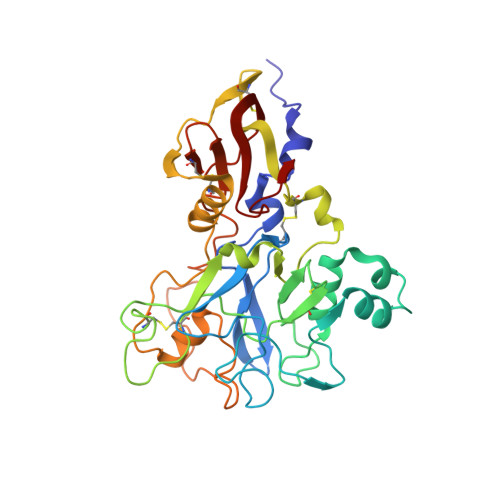
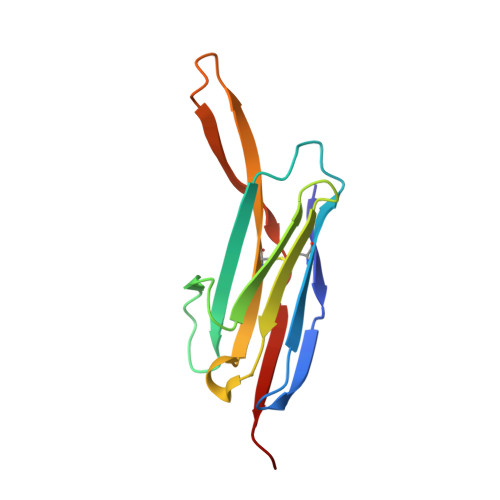
Structure of an IgNAR-AMA1 Complex: Targeting a Conserved Hydrophobic Cleft Broadens Malarial Strain Recognition
Henderson, K.A., Streltsov, V.A., Coley, A.M., Dolezal, O., Hudson, P.J., Batchelor, A.H., Gupta, A., Bai, T., Murphy, V.J., Anders, R.F., Foley, M., Nuttall, S.D.(2007) Structure 15: 1452-1466
- PubMed: 17997971
- DOI: https://doi.org/10.1016/j.str.2007.09.011
- Primary Citation of Related Structures:
2Z8V, 2Z8W - PubMed Abstract:
Apical membrane antigen 1 (AMA1) is essential for invasion of erythrocytes and hepatocytes by Plasmodium parasites and is a leading malarial vaccine candidate. Although conventional antibodies to AMA1 can prevent such invasion, extensive polymorphisms within surface-exposed loops may limit the ability of these AMA1-induced antibodies to protect against all parasite genotypes. Using an AMA1-specific IgNAR single-variable-domain antibody, we performed targeted mutagenesis and selection against AMA1 from three P. falciparum strains. We present cocrystal structures of two antibody-AMA1 complexes which reveal extended IgNAR CDR3 loops penetrating deep into a hydrophobic cleft on the antigen surface and contacting residues conserved across parasite species. Comparison of a series of affinity-enhancing mutations allowed dissection of their relative contributions to binding kinetics and correlation with inhibition of erythrocyte invasion. These findings provide insights into mechanisms of single-domain antibody binding, and may enable design of reagents targeting otherwise cryptic epitopes in pathogen antigens.
- CSIRO Molecular and Health Technologies, 343 Royal Parade, Parkville 3052, Australia.
Organizational Affiliation: