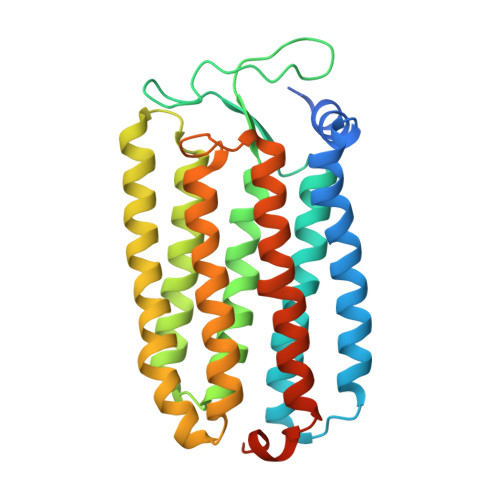
Crystal Structure of the Light-Driven Chloride Pump Halorhodopsin from Natronomonas pharaonis.
Kouyama, T., Kanada, S., Takeguchi, Y., Narusawa, A., Murakami, M., Ihara, K.(2010) J Mol Biology 396: 564-579
- PubMed: 19961859
- DOI: https://doi.org/10.1016/j.jmb.2009.11.061
- Primary Citation of Related Structures:
3A7K - PubMed Abstract:
The light-driven chloride pump halorhodopsin from Natronomonas pharaonis (phR) crystallised into the monoclinic space group C2, with a phR trimer per the asymmetric unit. Diffraction data at 2.0-A resolution showed that the carotenoid bacterioruberin binds to crevices between adjacent protein subunits in the trimeric assembly. Besides seven transmembrane helices (A to G) that characterise archaeal rhodopsins, the phR protomer possesses an amphipathic alpha-helix (A') at the N-terminus. This helix, together with a long loop between helices B and C, forms a hydrophobic cap that covers the extracellular surface and prevents a rapid ion exchange between the active centre and the extracellular medium. The retinal bound to Lys256 in helix G takes on an all-trans configuration with the Schiff base being hydrogen-bonded to a water molecule. The Schiff base also interacts with Asp252 and a chloride ion, the latter being fixed by two polar groups (Thr126 and Ser130) in helix C. In the anion uptake pathway, four ionisable residues (Arg123, Glu234, Arg176 and His100) and seven water molecules are aligned to form a long hydrogen-bonding network. Conversely, the cytoplasmic half is filled mostly by hydrophobic residues, forming a large energetic barrier against the transport of anion. The height of this barrier would be lowered substantially if the cytoplasmic half functions as a proton/HCl antiporter. Interestingly, there is a long cavity extending from the main-chain carbonyl of Lys256 to Thr71 in helix B. This cavity, which is commonly seen in halobacterial light-driven proton pumps, is one possible pathway that is utilised for a water-mediated proton transfer from the cytoplasmic medium to the anion, which is relocated to the cytoplasmic channel during the photocycle.
- Department of Physics, Graduate School of Science, Nagoya University, Chikusa-ku, Nagoya 464-8602, Japan. kouyama@bio.phys.nagoya-u.ac.jp
Organizational Affiliation: