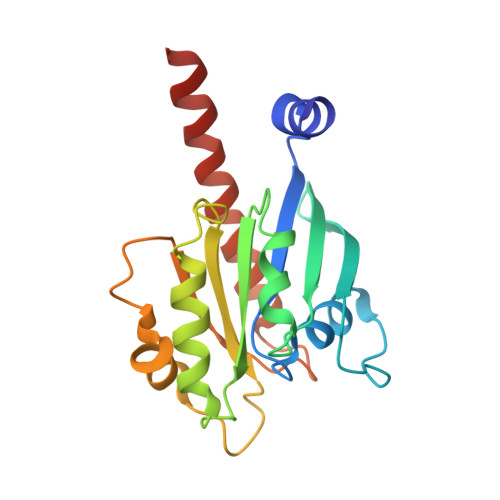
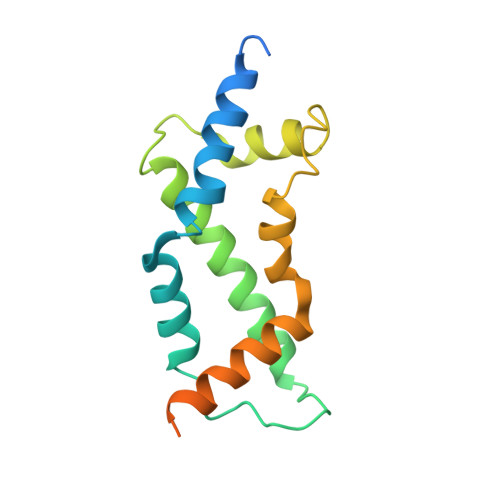
Crystal structure of the ARL2-GTP-BART complex reveals a novel recognition and binding mode of small GTPase with effector
Zhang, T., Li, S., Zhang, Y., Zhong, C., Lai, Z., Ding, J.(2009) Structure 17: 602-610
- PubMed: 19368893
- DOI: https://doi.org/10.1016/j.str.2009.01.014
- Primary Citation of Related Structures:
3DOE, 3DOF - PubMed Abstract:
ARL2 is a member of the ADP-ribosylation factor family but has unique biochemical features. BART is an effector of ARL2 that is essential for nuclear retention of STAT3 and may also be involved in mitochondria transport and apoptosis. Here we report the crystal structure and biochemical characterization of human ARL2-GTP-BART complex. ARL2-GTP assumes a typical small GTPase fold with a unique N-terminal alpha helix conformation. BART consists of a six alpha helix bundle. The interactions between ARL2 and BART involve two interfaces: a conserved N-terminal LLXIL motif of ARL2 is embedded in a hydrophobic cleft of BART and the switch regions of ARL2 interact with helix alpha3 of BART. Both interfaces are essential for the binding as verified by mutagenesis study. This novel recognition and binding mode is different from that of other small GTPase-effector interactions and provides molecular basis for the high specificity of ARL2 for BART.
- Institute of Biochemistry and Cell Biology, Shanghai Institutes for Biological Sciences, Shanghai 200031, China.
Organizational Affiliation: